Fig. 2
- ID
- ZDB-FIG-180221-38
- Publication
- Zhao et al., 2018 - Analysis of two transcript isoforms of vacuolar ATPase subunit H in mouse and zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
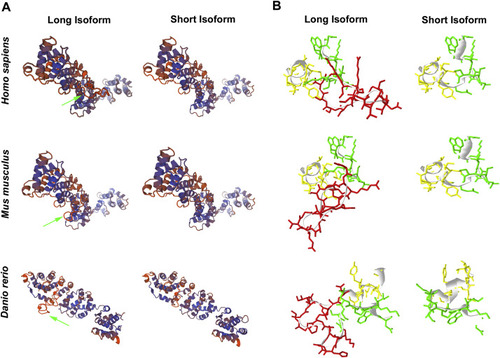
Comparison of three-dimensional structure using SWISS-MODEL analysis. A. 3D view of ATP6V1H protein. SWISS-MODEL 1ho8 and 3j9t are used to analysis ATP6V1H protein of Homo sapiens, Mus musculus and Danio rerio, respectively. The protein shows a large N terminal head and a small C terminal tail. Both N terminal and C terminal domain consist of some consecutive helices, and their connection presents as a shallow groove. The inserting eighteen amino acids in long isoform (green arrow) locate close to the central position of groove. Without the eighteen amino acids, the short isoform shows a shallower space between N part and C part, and the protein becomes looser. B. Local view of the inserting eighteen amino acids using Deepview. The eighteen amino acids are labeled in red and the ten amino acids before and after this domain are labeled in green and yellow, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) |
Reprinted from Gene, 638, Zhao, W., Zhang, Y., Yang, S., Hao, Y., Wang, Z., Duan, X., Analysis of two transcript isoforms of vacuolar ATPase subunit H in mouse and zebrafish, 66-75, Copyright (2018) with permission from Elsevier. Full text @ Gene