Fig. S3
- ID
- ZDB-FIG-161227-7
- Publication
- Frisca et al., 2016 - Role of ectonucleotide pyrophosphatase/phosphodiesterase 2 in the midline axis formation of zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
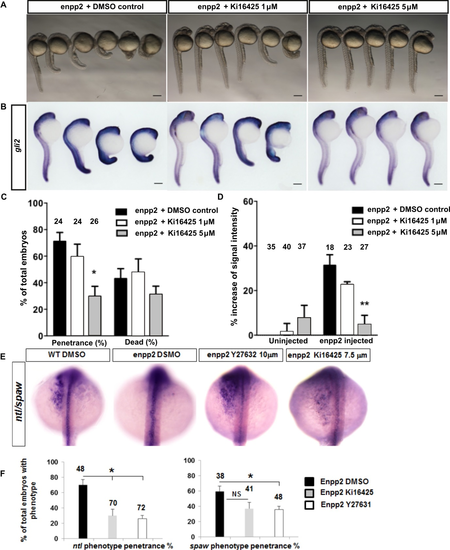
Antagonizing lpa1-3 using Ki16425 rescues the enpp2-overexpressed phenotypes. Representative bright field (A) and WISH images with gli2 riboprobes (B) of rescued experiments using Ki16425; in control (immersed in vehicle), Ki16425 (1 and 5μM) at 24hpf. The phenotype penetrance following enpp2 injection and Ki16425 treatment were measured at 24hpf (C, D). Following enpp2 overexpression, the phenotype observed was 71.1 ± 3.6 % in the vehicle control, 59.5 ± 9.5 % in Ki16425 1 μM, and 29.8 ± 7.5 % in Ki16425 5 μM. (C, D). E-F. Rescue experiments in enpp2 overexpressing embryos with Ki16425and Y27632 using ntl and spaw WISH as readout at 24 hpf. Following enpp2 overexpression, the ntl phenotype (notochord defects) was 70 ± 7.1 % in the vehicle control, 30± 8.5 % in Ki16425 7.5 μM, and 26 ± 4.3 % in Y27632 10 μM. spaw is normally expressed on the left side of the midline. The spaw expression phenotype (Figure 2 C, no expression, bilateral expression or expression on right side of the midline) was rescued in 36 ± 4.3 % of the Y27632 treated enpp2 overexpressing embryos. There was no significant rescue of spaw expression in Ki16425 treated embryos, although a clear trend was detected. Total number of embryos analysed stated above bar. Total number of embryos pooled from two independent experiments. Data are mean ± SEM. Statistical analysis was established by one-way ANOVA; * P < 0.05; ** P < 0.01. Scale bars: 200 μM. |