Fig. S3
|
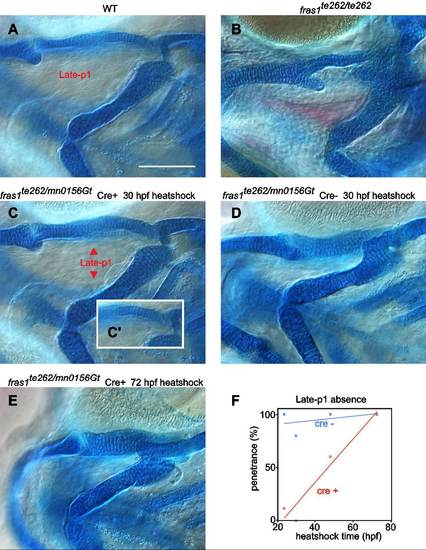
Cartilage and late-p1 phenotypes largely correlate in critical window experiments. (A–B) Differential interference contrast images of alcian/alizarin stained tissues of 6 dpf WT (A) and fras1te262 mutant homozygotes (B). (A) In WT, skeleton is well formed, and a completely outpocketed late-p1 is apparent. (B) In fras1te262 homozygotes, cartilage defects are seen and late-p1 is absent, as evidenced by mesenchyme occupying the space between ceratohyal and more dorsal skeleton. (C–E) 6 dpf alcian/alizarin skeletal preparations of fras1 reversion experiments, from crosses described in Fig. 7. Briefly, embryos trans-heterozygous for fras1mn0156Gt and fras1te262d were heat-shocked at different stages to induce Cre recombinase expression in the half of the clutch that inherited the hsp70l:Cre transgene; this Cre induction restores fras1mn0156Gt function. (C) If Cre is activated before the fras1 critical window (30 hpf), larvae form normal endodermal and skeletal morphology. (C’) Inset is a different focal plane showing normal symplectic morphology. (D) At 30 hpf, however, heat shock of control Cre siblings does not result in phenotypic rescue. (E) When heat-shock of Cre restores fras1mn0156Gt function after the critical window for fras1 function (72 hpf), both endodermal and skeletal phenotypes resemble fras1te262 homozygotes. (F) Plot of late-p1 defect penetrance as a function of time of development, determined by DIC imaging on one side of each embryo (n′s range from 5 to 13 for each condition). In Cre+ fish, there is a significant correlation between heat shock time and late-p1 defect penetrance (R2=0.92, P<0.05), however there is no significant correlation in Cre- fish (R2=0.17, P=0.6). Scale bar is 100 µm, applicable to A-E. |
Reprinted from Developmental Biology, 416(1), Talbot, J.C., Nichols, J.T., Yan, Y.L., Leonard, I.F., BreMiller, R.A., Amacher, S.L., Postlethwait, J.H., Kimmel, C.B., Pharyngeal morphogenesis requires fras1-itga8- dependent epithelial-mesenchymal interaction, 136-48, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.