Fig. S2
- ID
- ZDB-FIG-160303-39
- Publication
- Stegmaier et al., 2016 - Real-Time Three-Dimensional Cell Segmentation in Large-Scale Microscopy Data of Developing Embryos
- Other Figures
- All Figure Page
- Back to All Figure Page
|
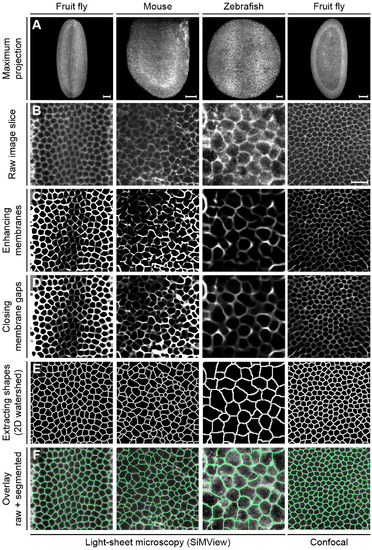
Related to Figure 1. Fast and accurate slice-based 2D segmentation Illustration of the main processing steps performed prior to fusion of the 2D segments. Maximum intensity projections of 3D image stacks of Drosophila, mouse and zebrafish embryos recorded with SiMView light-sheet microscopy and confocal fluorescence microscopy are shown in panel (A). Based on the raw membrane image (B, xy-image is shown, i.e. an image slice oriented perpendicular to the microscope’s optical detection axis), a Hessian-based objectness filter is used to enhance membrane structures (C). Remaining holes in the cell membranes are repaired using iterative morphological closing with increasing structure element sizes (D). The actual segmentation is performed using a standard watershed algorithm on the 2D slices (E). The last row (F) shows the watershed pixels superimposed with the raw images. Image contrast has been adjusted for better visibility. Scale bars, 50 µm (A), 20 µm (B). |
Reprinted from Developmental Cell, 36, Stegmaier, J., Amat, F., Lemon, W.C., McDole, K., Wan, Y., Teodoro, G., Mikut, R., Keller, P.J., Real-Time Three-Dimensional Cell Segmentation in Large-Scale Microscopy Data of Developing Embryos, 225-240, Copyright (2016) with permission from Elsevier. Full text @ Dev. Cell