Fig. 7
- ID
- ZDB-FIG-150324-18
- Publication
- Liu et al., 2015 - Molecular evolution and functional divergence of zebrafish (Danio rerio) cryptochrome genes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
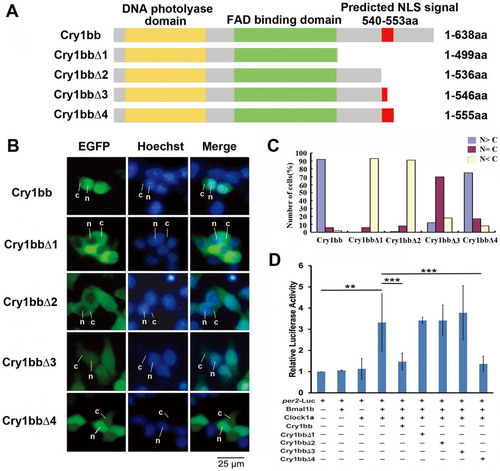
Identification of a novel NLS (Nuclear Localization Signal) sequence of zebrafish Cry1bb. (A) Schematic diagrams of the full length of zebrafish Cry1bb (1–638aa) and Cry1bb truncation constructs. The yellow bar indicates the DNA photolyase domain, the green bar indicates the FAD binding domain of Cry proteins and the red bar indicates the predicted NLS sequence (VRREQPGPSGAKHR, Ensembl Peptide ID: ENSDARP00000090995) according to an online program (www.moseslab.csb.utoronto.ca/NLStradamus)46 (see also Supplementary Fig. S7). Cry1bbΔ1 contained 499amino acids, truncated at amino acid position number 499; Cry1bbΔ2 contained 536amino acids truncated at amino acid position number 536 just before the predicted NLS signal; Cry1bbΔ3 contained 546amino acids truncated at amino acid position number 546 in the middle of the predicted NLS signal; and Cry1bbΔ4 contained 555amino acids truncated at amino acid position number 555 just after the predicted NLS signal. (B) Sub-cellular localization of zebrafish Cry1bb and its four truncated fragments. Cry1bb-EGFP or Cry1bb-EGFP truncated fragments were transfected into HEK293 cells, respectively, and then the cells were stained with Hoechst. The cytoplasmic or nuclear distributions of zebrafish Cry1bb and its four truncated fragments were detected by GFP signals (green); the nuclei were identified by Hoechst (blue). The leader lines and letters N, C indicate nuclear and cytoplasmic localizations, respectively. (C) Quantitative analysis of the subcellular localization of each Cry proteins. In each experiment, 50–100 cells were evaluated for nuclear fluorescence (N > C, blue bars), nuclear-cytoplasmic fluorescence (N = C, purple bars), and cytoplasmic fluorescence (N < C, pink bars). The figure shows a representative of the three independent experiments. (D) Repressive activities of zebrafish Cry1bb and its four truncated fragments determined by dual Luciferase reporter assays. The full length of Cry1bb and its four truncated fragments were co-transfected with per2-luc reporter and Bmal1b:Clock1a combination, respectively. Among the four Cry1bb truncated fragments, only Cry1bbΔ4 retained the repressive ability of the full length Cry1bb on Bmal1b:Clock1a-mediated transcription. A Renilla Luciferase was added in each transfection to normalize transfection efficiency. The figure shows the mean and SD (error bar) of two independent experiments (triplicate for each experiment). Results were analyzed by ANOVA. |