Fig. 6
|
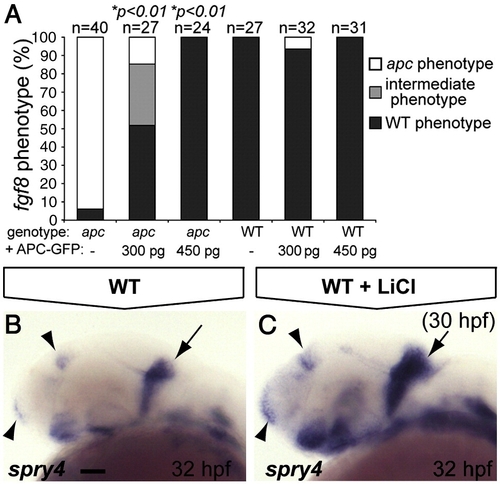
The IsO phenotype as revealed by fgf8 labeling is mainly the result of β-catenin stabilization. (A) Percentage of genotyped mutant, wild-type and heterozygous sibling embryos, showing mutant (white), intermediate (gray) or wild-type (black) phenotypes upon injection of 300 or 450 pg apc-GFP mRNA. The number of injected embryos is indicated above the bars. Injection of 450 pg apc-GFP mRNA completely rescues fgf8 patterning defects. (B, C) LiCl treatments at 30 hpf induces expanded expression at the MHB (arrow), telencephalon and diencephalon (arrowheads) of the Fgf target gene spry4 (C). Lateral view. Anterior to the left. Statistics: two-tailed Fisher′s exact probability test for a table of frequency data. p-values are indicated above the bars. Scale bar 50 μm. |
| Gene: | |
|---|---|
| Fish: | |
| Condition: | |
| Anatomical Terms: | |
| Stage: | Prim-15 |
Reprinted from Developmental Biology, 331(2), Paridaen, J.T., Danesin, C., Elas, A.T., van de Water, S., Houart, C., and Zivkovic, D., Apc1 is required for maintenance of local brain organizers and dorsal midbrain survival, 101-112, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.