Tech. Report CIS-TR-97-02, Computer and Information Science Dept., University of Oregon, 1997.
University of Oregon
Eugene, OR 97403
1.0 INTRODUCTION
The technology of the World Wide Web (WWW) provides a revolutionary means for dissemination of scientific information. For the first time, scientists have 24 hour, low-cost international access to central repositories of research data without the need for specialized client-side software. In particular, biological researchers have exploited the power of the Web to create a diverse range of bioscience resources, including several web-accessible relational databases (e.g., Mouse Genome Database [7], the Human Genome Database [8], the C. Elegans database [3], the Genome Sequence Database [10], and FlyBase [6] ). There is also a growing number of other Web sites serving static HTML documents; by spring of 1996 there were 26 different Web sites for 15 different species, and the number of sites is increasing at a rate of about one every three to four months.
The biologists and computer scientists constructing these web sites presume that these web-accessible resources will aid scientific discovery through more timely, widespread access to better integrated research information. Although the WWW has made this information physically accessible to scientists, it is unclear whether it will be cognitively accessible. Busy scientists want useful, accurate, complete and up-to-date information without needing to learn and use a complex user interface. Will they be able to find answers to their questions without resorting to powerful but complex query languages like SQL? Will they be able to get their answers quickly without working through endless hierarchies of useless pages? Accessibility depends upon usability, and usability is critically related to productivity [9].
Designing a usable interface is challenging. First, the interface design must observe sound principles of graphic arts and psychology; it must have a functional layout, recognizable icons, and consistent interaction styles. Good design will reduce the time required to access desired information by minimizing misconceptions, mistakes and confusion in the search process. Second, it must incorporate a deep understanding of what information the scientist needs in the immediate context of his or her tasks and activities. In other words, it must present information using language and conceptual models understood by the scientist.
We have created a Web-based biological database for the zebrafish research community. The success of our project and the achievement of true accessibility and productivity, depend upon designing, developing, and implementing with a user-centered approach. In taking this approach, we believe we are relatively unique among designers of web-accessible data resources. Here, we describe our use of this method, the specific user interface challenges we faced, and the resulting design.
2.0 OVERVIEW OF THE ZEBRAFISH INFORMATION NETWORK (ZFIN) PROJECT
Researchers using zebrafish to study basic biology, like genetics and development, are distributed among more than 100 laboratories in 28 countries. The zebrafish database project evolved out of our earlier Web site [16], which makes available (in static HTML documents) information on researchers and labs, a bibliography of publications relevant to the zebrafish research community, photos illustrating zebrafish developmental stages, and descriptions of laboratory methods, mutant lines, and the genetic map. The home page was accessed over 20,000 times in its first year.
Due to the exponential increase in information in this research area and the resulting need for more powerful methods of organizing and accessing these data, the zebrafish research community mandated extension of the original Web server to create a WWW accessible multimedia relational database known as the Zebrafish Information Network (ZFIN).
3.0 THE USER-CENTERED DESIGN : PROCESS
User-centered design focuses on the ultimate usefulness and usability of an interactive software product by assessing the requirements and specifications of the product from the user's point of view, the user's interactive behavior when using the software, and the context of its use. Our design process (Figure 1) follows the basic steps of user-centered design [15].
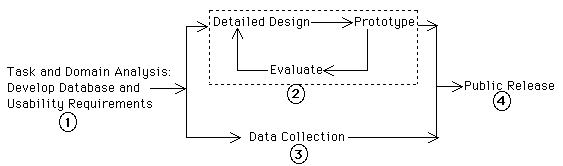
Step 1: Develop database and usability requirements
The initial step in our design process is to conduct domain and task analyses. The goal of this step is to produce both database information and interactive system specifications. However, the biological domain is extremely specialized, making this analysis very difficult. Development of the abstract data model, the nomenclature used to label user interface components, and the structuring of interface actions into information seeking tasks all require deep knowledge of the domain to achieve efficiency and usability. Accordingly, we began the design process by forging a participatory design team [4, 15] which includes both biologists and computer scientists. We believe that this collaboration is key to the success of our project, not only because it provides domain and task knowledge, but also because direct involvement of biologists gives them a stake in the success of the project.
During Step 1 (Figure 1) we used primarily ethnographic methods [1], including interviews with zebrafish scientists, reading journal articles, attending research talks and lab meetings, and participant-observer activities such as helping to customize the specialized software used by one of the labs. In other words we tried to "go native" in the zebrafish community. We also used questionnaires to gather design input from scientists around the world, distributing them at workshops, and via the original Zebrafish Web site, which contains documents on the development of the database project along with a brief list of the types of information we expect to include. We solicited feedback from our users about their satisfaction with the current HTML-based Web site and integrated their requests for enhanced functionality and information into the design of the new database. The goal of this immersion in the working world of zebrafish scientists was to understand the context of their everyday work activities and its relationship to the proposed WWW database.
In addition to these ethnographic methods, we looked extensively at other web-accessible biological database sites to evaluate their information content and user interfaces; we videotaped our own zebrafish scientists doing simple information retrieval tasks to pinpoint their confusions with the user interfaces and information models at these other sites. In this respect, we have found the WWW to be an easily accessible means of drawing on the design experience of others, a critical resource in an area where design improvements are typically incremental and based on real-world experience.
These domain and task analyses produced specifications for the database information and the user interface. Database information specifications were captured in a data model document intelligible to both computer scientists and biologists, containing descriptions of database entities, attributes, and relationships, as well as examples of situations of use for various pieces of information. The data model serves as the blueprint for database implementation and offers zebrafish biologists a concise overview of database contents. The current design for the database is complex and incorporates 21 major classes of information, most of which are highly interconnected.
Step 1 (Figure 1) also produced specifications for the interactive system. In particular, functional and performance requirements of the system were determined as seen from the user's point of view [12]. The first of these, functional requirements, describes what the system should do. Our functional requirements include:
- Must provide a security mechanism
to ensure that only authorized users submit data.
- Must guarantee reliability and completeness
of the data, especially because much of it is user supplied.
- Must have a mechanism to distinguish
published data from unpublished or pre-published data.
- Must allow submission of commonly
published image types, including annotations.
- Must provide color reliability and
reasonable resolution for images so that information is accurate
enough to make science-based decisions.
- Must be accessible from multiple
platforms, including older machines, to support universal
access to the database.
- May need to access other databases concurrently with ZFIN.
Performance requirements, the second type of requirements, state
how well the system should perform from the user's point of view.
Thus, performance requirements define criteria for evaluating
the actual usability of the resulting design used in Step 2 (Figure 1) of the design process. Our performance requirements include:
- Must be easy to learn. We expect
user interactions with the database (retrievals or submissions)
to be relatively infrequent, and thus, a typical scientist might
forget how to use the interface between uses. The interface must
be learned as you go (no separate manual) and must provide extensive
on-line help.
- Must be fast enough to satisfy most
commonly asked questions within a 10 minute session.
- Must provide enough feedback that
most searches will find results with three rounds of querying.
- Must keep the user apprised of progress during the data submission process, allowing the user to "undo" a submission during any step.
It was apparent from our requirements study that we needed to offer a more usable interface than SQL. Although SQL is extremely expressive, allowing extraction of very complex database relations, it is practically impossible for non-database professionals to learn and use [5]. Thus, a primary challenge in the design of the ZFIN user interface was to determine in advance a subset of queries which would satisfy the needs of most users and to create a very simple interface for expressing queries in this subset. This required an extensive understanding of the domain and tasks.
Constraints Imposed by HTTP/HTML
The functional and performance requirements listed above derive from the characteristics and tasks of the zebrafish research community. Interface design for any Web-accessible application is also severely constrained by a number of basic limitations in the technology of the Web itself, primarily because the WWW environment was designed to support distribution of static multimedia documents, rather than dynamic connections between clients and server-side applications. This bias is reflected in Hypertext Markup Language (HTML), the simple formatting language used to control interface layout, and in Hypertext Transfer Protocol (HTTP), the communication protocol used to implement client-server interaction. By limiting the interface primitives available to the designer and the ways in which client and server can interact, HTML and HTTP impose a number of unusual constraints on interface design:
-
Inconsistent Look and Feel. A basic premise of HTML is
that the language should logically describe the appearance
of interface screens, relegating to the client software low-level
decisions associated with physically displaying the page. Despite
recent improvements, control over the layout and appearance of
interface elements remains relatively weak. Consequently, appearance
of the interface may vary widely depending upon the client hardware
platform and browser software used to access the interface.
Discrete Transaction Model. The HTTP protocol supports only discrete transactions between client and server. Each transaction retrieves a single HTML document. Transactions may be initiated only from the client side; the server cannot spontaneously send information to the client.
Anonymous, Stateless Transactions. Both HTTP server and client treat each transaction as completely independent of preceding transactions. Though both typically maintain some record of document access, this record plays no role in shaping the retrieval or appearance of future documents. Moreover, the HTTP protocol does not provide integrated mechanisms for distinguishing between transactions at the user (client) level, meaning that transactions are essentially anonymous.
The functional requirements and performance requirements combine with the constraints imposed by HTML and HTTP to drive the interface design.
Step 2: Iterate detailed design process
Step 2 (Figure 1) is the heart of usability engineering methods [13]. After developing the information and usability requirements, we moved into the iterative refinement phase of the user-centered design process to design and implement the user interface. In contrast to the waterfall approach used by traditional software engineers, this technique relies on rapid cycles of design, prototype implementation, and evaluation with real users to generate the final product.
Rather than implementing the entire database at once, we initially selected a subset of the database information (i.e. data types) to take through Steps 2-4 of the design process. This was done primarily for pragmatic reasons. Some information is of higher priority to the research community or more mature and complete; staggering development of various data classes allowed us to make useful information rapidly available. In addition, focusing attention on just a few types of information at a time proved to be an effective means of managing the complexity of the design.
Each prototype was immediately evaluated by selecting pairs of zebrafish scientists and giving them typical data retrieval and submission tasks to perform. Videotaping these sessions allowed us to analyze the amount of time required, misconceptions encountered, and other problems with the interface. We evaluated their performance against the usability requirements developed in Step 1 (Figure 1). Details of how to conduct this type of performance analysis can be found in [2]. We used insights gained from this analysis to shape subsequent prototypes in the iterative design cycle.
When we were satisfied with the usability of a prototype, it was made available to 10 zebrafish scientists (acting as beta testers) through the WWW; access to the prototypes was limited to these testers. A "comment form" was integrated into each screen, allowing our testers to record feedback easily and email it to the developers. At the end of the beta testing period, we also interviewed our testers to discover any other problems.
Steps 3 and 4: Data Collection and Public Release
Because data collection (Step 3) is not an intrinsic part of the user-centered design process, it will not be addressed here, in favor of a more focused discussion of usability and interface design issues. Step 4, public release of the database, is an important part of the user-centered design process rather than its abrupt termination; usability analysis will continue indefinitely, allowing the system to evolve to meet the changing demands of users. The commentary forms for gathering user feedback mentioned earlier in the context of beta testing remain available in the public release. We are also recording (anonymously) the sequence of screens visited by each user and the total number of visits to each screen in an effort to determine common usage patterns and to expose areas of confusion. Finally, we are planning to conduct extensive periodic user surveys, interviewing a sample of randomly selected registered ZFIN users to assess usage patterns, good and bad features of the Web site, and interest in future information support.
4.0 USER-CENTERED DESIGN: PRODUCT
4.1 User Interface Architecture
Although several research efforts have produced interfaces to interactive server-side applications that rely solely on the mechanisms available in HTML/HTTP [14], we found it impossible to satisfy our demanding usability requirements within these constraints. Therefore, our implementation integrated three powerful concepts to enhance significantly the expressive power of the ZFIN interface:
-
Interface state. The ZFIN architecture takes advantage of
the underlying database to store and maintain information about
users and interface state. In combination with user authentication,
this provides strong support for data security, navigation and
context-sensitive help.
Personalized interaction. By providing mechanisms for authenticating and identifying individual users, the ZFIN interface tailors its presentation to meet the specific needs of each user.
Client-side mechanisms. Responsiveness and feedback of the interface is enhanced by taking advantage of client-side processing wherever possible, distributing responsibility for interface management more evenly between client and server.
We implemented these concepts in a hybrid processing architecture that augments conventional HTML specification with the storage capabilities of the underlying database, the computational and formatting capabilities of a powerful scripting language, and the interactivity enhancements of client-side processing (e.g. Java and Javascript) to create a flexible, reactive interface programming environment.
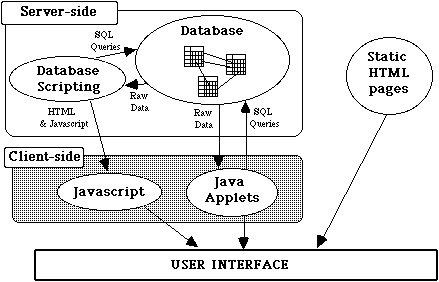
Four distinct mechanisms are transparently integrated to create the user interface (Figure 2): a relational database, dynamically generated HTML pages, Java applets and static HTML pages. Static HTML pages are the most trivial component and warrant no further discussion, especially because they are only rarely used in our dynamic interface (e.g. for displaying introductory descriptions).
Dynamic page generation, shown on the left in Figure 2, creates the majority of interface screens in ZFIN. Raw "application pages", consisting of static elements interspersed with scripted processing instructions, are stored as text in a database table. We used two distinct types of scripted instructions: database scripts, which control the content and appearance of the returned page, and Javascript, which controls client-side dynamics like checking input for errors, enforcing semantic constraints, and updating the navigation display (discussed in upcoming sections).
As an application page is retrieved from the database, it undergoes a sequence of transformations to generate the final interface screen presented to the user. First, the database scripts embedded in the page are executed, issuing SQL statements to the database server and using the returned values to control page layout and to populate the nascent HTML page with formatted data. These scripts may also generate and embed page-specific Javascript instructions to control dynamic client-side behavior. Variables passed in the original page request are also available to the script interpreter, and may shape the returned page. The resulting HTML page is passed back to the user's browser, which executes any embedded Javascript instructions and displays the final product.
Although this layering of scripting mechanisms greatly enhances flexibility, the end product is still HTML, which does not support dragging, pointing and other forms of direct manipulation. For some tasks, there was simply no way to meet the usability criteria developed Step 1 (Figure 1) of our design process without access to these more advanced interaction techniques. Thus, the architecture also allows the integration of special purpose Java applets1 to implement advanced interface functionality.
The only cost incurred by this architecture is a slight decrease in accessibility; the integration of client-side technologies like Java and Javascript means that, to access the interface, users must have browsers that support these technologies.
4.2 User Interaction: The ZFIN Interface
The interface supports four primary interaction modes (Figure 3): surveying database contents and activities, querying to retrieve information, submitting and updating data, and using specialized data manipulation tools.
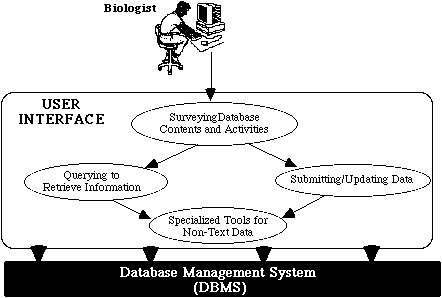
4.2.1 Surveying Database Contents and Activities
Our analysis of usability requirements during Step 1 of the design process showed an overwhelming demand for immediately productive, efficient access to information for untrained users.
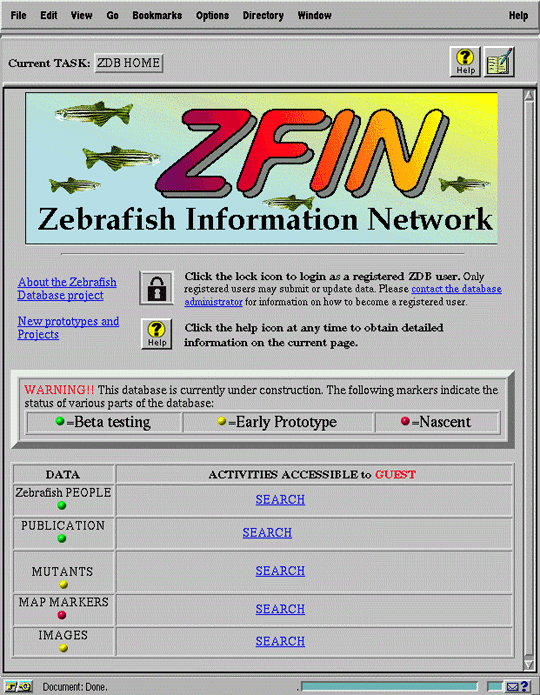
The ZFIN "home page" (Figure 4a) meets this requirement by immediately presenting users with an overview of available data and accessible activities. In this way, the interface establishes a simple conceptual model of the data space and streamlines the most common access scenario, namely, performing a basic search for information. The "status" of the data in each category is indicated, priming users' expectations about data integrity and completeness.
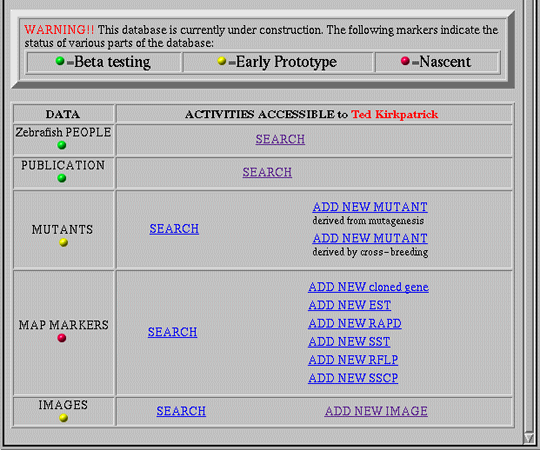
Figure 4 also illustrates how the concept of personalized interaction [17] simplifies the interface. By distinguishing among users, both as individuals and as members of pre-defined groups, the interface presents each user with a customized perspective that contains only those elements accessible to the user. In this way, the interface focuses the user's attention on relevant activities, while concealing unnecessary complexity. For example, when users access ZFIN, they are initially classified as "guests", permitting them free viewing access to all data, but barring them from submitting updates or new data. The latter activities are reserved only for "data submitters", scientists within the zebrafish research community who have applied for and received authorization to submit data. Figure 4b shows the lower portion of the home page seen by Ted, a member of the "data submitter" group, after he has logged in. As a data submitter, Ted has access to a set of activities centered around data submission which are unavailable to guest users (Figure 4a). Note, however, that Ted is unable to add new persons or publications to the database; these activities are reserved for the unrestricted "database administrator" group, of which Ted is not a member.
We implemented identification of individual users by using the "HTTP cookie" mechanism [11] in combination with a table of user information stored in the underlying database. When a user logs in, name and password are checked against the user table and, if successful, a randomly generated "identity key" is installed as an HTTP cookie in the user's browser; the key is also associated with the user's record in the database. With every subsequent transaction, the user's browser passes the cookie to ZFIN to establish identity and permission status. This interface then uses this information during the dynamic page-generation process to determine whether the user has permission to access a requested page and, if so, which activities to make accessible to the user within the page.
The ZFIN home page (Figure 4a) also establishes the overall layout of the interface, with navigation tools, help, and lab notebook available at the top of each screen, and a "content" area below. This structure is consistently maintained throughout subsequent screens.
4.2.2 Querying to Retrieve Information
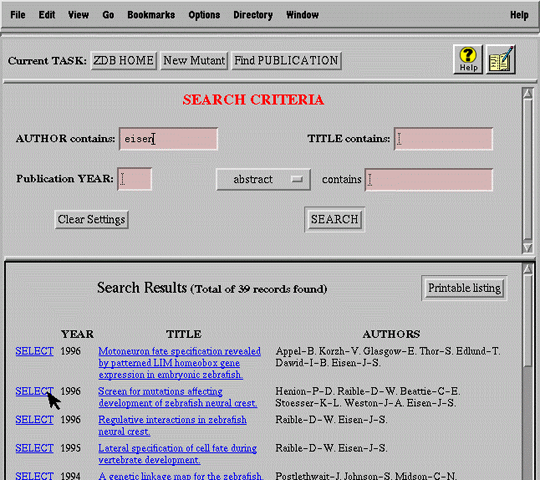
Focused search is very different from the more general browsing activity supported by traditional WWW sites; users typically have a well-defined set of target criteria in mind when they initiate the search. A successful interface must allow efficient expression of these criteria using domain terminology, must summarize search results, and must maintain the connection between search criteria and matching records.
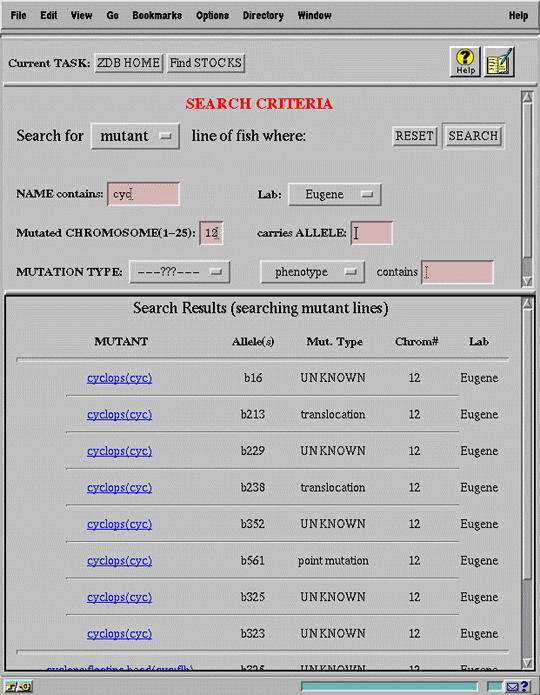
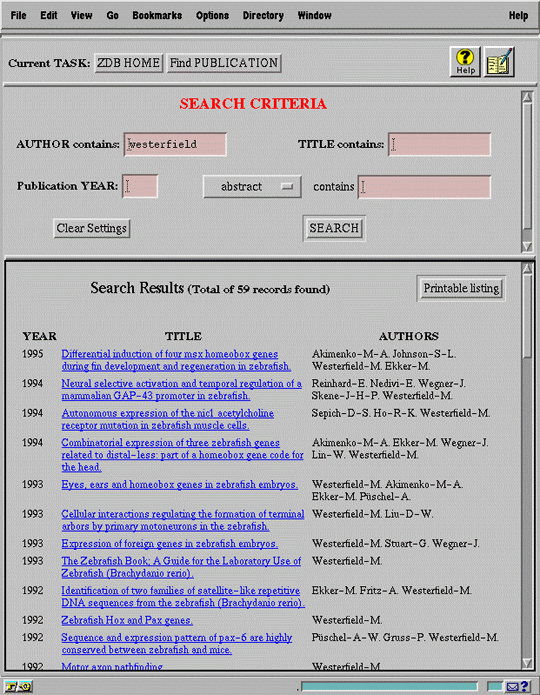
Figure 5 presents the search interface implemented in ZFIN. The simple layout and limited number of search criteria reflect our commitment to accessibility and ease of use rather than expressive power. Although the underlying database query language (SQL) allows formulation of arbitrarily complex queries, the interface limits constraints to conjunctions of a handful of criteria identified by our domain analysis as most useful (i.e. most discriminating, most likely to be known at time of search).
The interface maintains the conceptual connection between search criteria and search results by juxtaposing them on the same screen; there can be no confusion about the criteria that produced the currently displayed set of records. More generally, the spatial relationship between criteria and results visually relates the user's goals and actions to the responses produced by the system. This arrangement also makes iterative refinement of the search very easy, applying new constraints while observing the effect on the set of matching records.
The two screens shown in Figure 5 also illustrate the consistency of the search interface between data classes. Although the criteria are different for each class, the basic screen layout and search process are identical.
4.2.3 Data Submission and Updating.
Support for user submission and updating of experimental data is one of the most important and unique features of the ZFIN interface. We encountered four primary challenges during our design of the data submission interface: supporting naive users, enforcing data security, supporting multi-step submissions, and providing context-sensitive navigation and help.
Supporting naive users.
To support inexperienced users, the data submission interface
provides instantaneous feedback wherever possible, checking entries
for correct format and enforcing semantic constraints between
fields. For example, error dialogs appear when values are out
of range, incorrectly formatted, or incompatible with previously
specified values. We implemented this feature by embedding context-specific
Javascript instructions in each data submission screen.
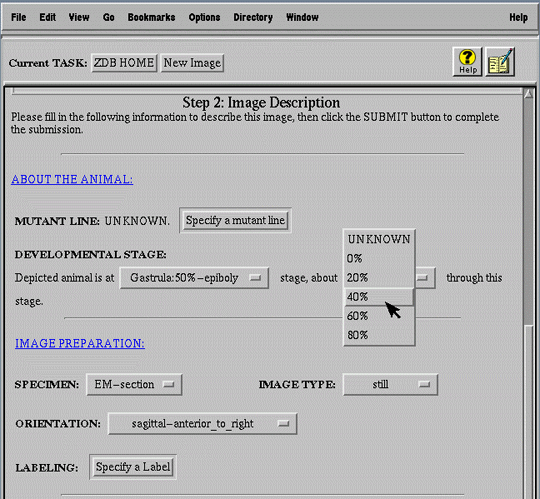
Another important ease of use heuristic employed in the interface is the preference for popup menus over text entry (Figure 6a). Popup menus are particularly helpful to infrequent and inexperienced users who may not remember or know the possible values or valid data formats for each field. They also provide a convenient means of enforcing uniform biological nomenclature, increasing the effectiveness of data indexing and retrieval.
Enforcing data security .
In any network environment, allowing remote data modification
greatly increases the risk to data security. Our user authentication
mechanism conveniently enforces data security, selectively restricting
access to allowed data manipulation activities. The interface
enforces security not only at the page level, but on a feature
by feature basis within each page. For example, the interface
allows the user to update his or her personal record (Figure 6b),
except name and publications; these options appear only
to the database administrator.
Supporting a nested, multi-page submission process.
The interdependence of experimental data presented a major challenge
in the design of the data submission interface. A "data submission"
includes not only new information, but also specifying the myriad
relationships between that datum and other records contained in
the database. The need to establish connections between a new
record and related records results in a complex, nested data submission
process in which the submitter must digress repeatedly from the
main submission task to search for and create links to related
information.
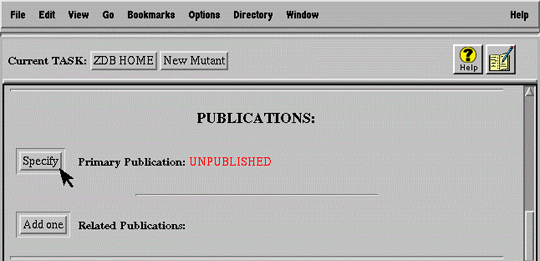
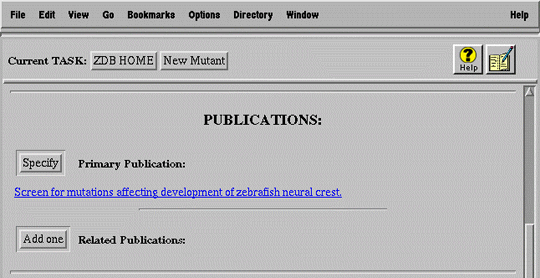
For example, a submitter must digress from the primary task of submitting a new mutation (Figure 7a) to search for the publication (Figure 7b) in which the mutation was first described (i.e. primary publication). After finding the publication, the user selects it and automatically returns to the mutant submission form (Figure 7c).
The basic HTTP/HTML environment cannot accommodate this complex, nested sequence of task and digressions (subtasks) because it supports no notion of interface state. The central difficulty lies in managing the automatic "return" from a subtask to the task from which it was initiated, bringing the information specified in the subtask back to the primary task.
We implemented this functionality by saving interface state2 in the underlying database and passing a reference (pointer) to that state to pages associated with the selected subtasks. Specifically, the record of interface state captures two important pieces of information as a digression is initiated: the values contained in the partially completed data submission form and the position (i.e. viewport) within that form to which the interface should return when the digression ends. As a first step in any digression, a "temporary" record is created in the database and filled with the current values of the form's input fields. The unique identifier for this "stack frame" is then passed along to subsequent pages as a hidden form element. When the digression ends, the values stored in the temporary record are retrieved and the original form reconstructed; from the user's point of view, the interface has "returned" to a previous screen.
Providing context-sensitive navigation and help.
Not all digressions are as straightforward as the one presented
in Figure 7. A submitter may need several tries to fine-tune search
criteria, or may need to view one or more records in the candidate
set before selecting one. This raises another prominent design
challenge: during "deep" digressions, users easily become
confused about the relevance of the current screen to the task
solution process as a whole. In other words, users tend to "get
lost" in the task space. Left unsupported, their only remedy
is to return to the home page and start over.
Initially, we assumed that the history mechanisms built into most browsers would adequately support navigation through the task space, allowing users to view and move back through a list of previously traversed pages. However, our usability testing revealed a crucial difference between casual browsing and task-oriented data access. In a task-solution process, a one-to-many (rather than a one-to-one) correspondence exists between the physical pages retrieved and the conceptual steps within the process. For example, iteration of the search-retrieve cycle (i.e. while refining constraints) generates a lengthy sequence of retrieved pages associated with the same conceptual step (i.e. find candidate data) in the search task. All of these pages appear in the browser's history list, leaving the user confused as to where they all came from (not realizing that each new search generated a new page) and how to get back to the preceding step in the task. What is needed is a task-sensitive mechanism that maintains a user's orientation within the task space by tracking his or her progress through the conceptual steps within each task.
The ZFIN navigation tool provides users with a dynamically-updated representation of their current position within the task space. Beginning with the "home page", tiles corresponding to each conceptual step in a task appear in the "control strip" found at the top of each page (see Figures 4-7) as the user embarks on that step. At any point within a digression, a click on the tile representing some earlier, enclosing step aborts the digression, instantly returning the user to the earlier step.
The explicit representation of the user's current task also supports a powerful context-sensitive help mechanism. Because the interface maintains a complete history of a user's interaction, it is able to provide specific guidance on how to complete the current task.
4.2.4 Specialized Tools
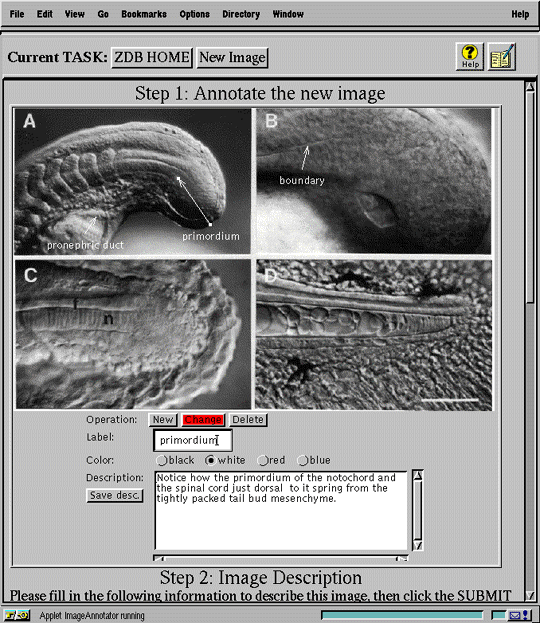
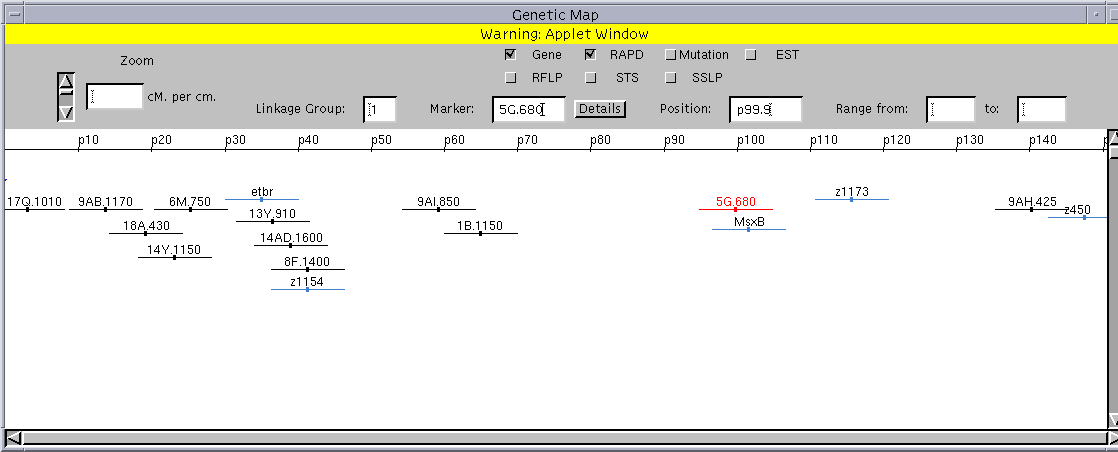
Although the interface features described in the preceding sections on searching and data submission are adequate for describing, retrieving, and displaying text data, they do not efficiently3 support manipulation of complex, domain-specific data types like annotated images and genetic maps.
We developed two Java applets to support these data types. The image annotator (Figure 8a) allows the user to place annotations dynamically on a submitted image. The genetic mapping engine (Figure 8b) is a sophisticated tool for analyzing the spatial relationships among genetic markers appearing on a selected segment of a chromosome. Both applets communicate directly with the database server to retrieve data required by the applet or, in the case of annotations, to add information to the image record. These specialized tools enhance usability by supporting directly the conceptualizations of domain data prevalent in the biological domain, transparently mapping them to underlying data representations.
5.0 CONCLUSIONS
Rapidly expanding access to the WWW holds incredible promise for increased data sharing and collaboration within widely-distributed research communities. Web-accessible databases will make available a much broader range of data than printed media, including multimedia data types and information that, although useful, might never be formally published; new findings can be made available almost instantly, rather than being delayed for months by a lengthy editorial and printing process.
There are a number of challenging obstacles to such universal accessibility. First, scientific domains are unusually complex, with domain ontologies that evolve with the expanding frontiers of the discipline; the kinds of information accepted as "data" and the research techniques that generate this information change over time. Consequently, interface design for a research database is much more demanding than for stable, single-use databases. From a practical perspective, the WWW environment imposes challenging constraints on the design on a web-accessible interface, severely limiting the interface concepts and interaction techniques available to the designer.
The case study presented here shows how user-centered design can be used to manage domain complexity and generate meaningful usability requirements, by focusing attention on the real-world work activities (i.e. research processes) of users and on the ways in which access to various kinds of information (data) contributes to these activities. It also demonstrates how the constraints imposed by the WWW environment can be greatly ameliorated by extending the rudimentary interaction model defined by HTTP/HTTP with three powerful concepts: interface state, personlized interaction, and client-side dynamics.
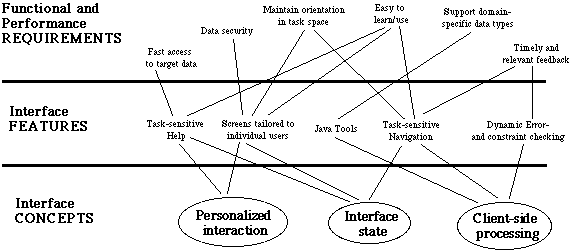
Figure 9 summarizes our observations on the relationships among these underlying interface concepts, the specific interface features they give rise to, and the ways in which these features support the usability criteria generated by our user-centered analysis.
At the implementation level, our work demonstrates that the integration of several technologies, an underlying database, HTML, database scripting, Javascript and Java, establishes a flexible substrate for creating expressive and reactive web-accessible interfaces. By judiciously interweaving these technologies to capitalize on their respective strengths, the ZFIN interface architecture defines a pragmatic balance between simplicity and expressive power.
We have focused our work to date on maximizing the accessibility of data contained in the ZFIN database, applying user-centered techniques to streamline individual data manipulations. In the future, we plan to expand our focus to support the full research process in which database access is embedded by developing tools to support database-centered interaction among widely-distributed members of the research community. Examples include mechanisms for collaborative shared access to the database, interactive discussion forums, and viewer commentary appended to specific data records. We envision ZFIN as the cornerstone of a virtual community of research scientists linked via the WWW, working together, and sharing a common set of data.
Acknowledgments
Mike McHorse and Paul Bloch provided essential system administration support for our computers and network. The base code for the genetic mapping applet was written by Gregg Helt and provided by the bioWidgets Consortium. We would also like to thank our numerous informants within the zebrafish research community, who patiently took time to explain their research methods to us. The zebrafish database project is sponsored by NSF grant BIR-9507401.
References
- Blomberg, J., Giacomi, J., Mosher, A., & Swenton-Wall, P. (1993). Ethnographic Field Methods and their Relation to Design. In Schuler, D., & Namioka, A. (Eds.). Participatory Design: Principles and Practices. Hillsdale, NJ: Lawrence Erlbaum Assoc.
- Douglas, S.A. (1995), Conversation Analysis and Human-Computer Interaction Design. In P.J. Thomas (Ed.) Social and Interactional Dimensions of Human-Computer Interfaces. Cambridge University Press, 1995, pp.184-203.
- Genome Informatics Group. (1996). ACEDB (World Wide Web URL http://probe.nalusda.gov:8300/cgi-bin/query?dbname=acedb). Beltsville, MD: US Department of Agriculture.
- Greenbaum, J., & Kyng, M. (1991). Design at Work: Cooperative Design of Compiuter Systems. Hillsdale, NJ: Lawrence Erlbaum Assoc.
- Greene, S.L., Gomez, L.M., & Devlin, S.J. (1986). A cognitive analysis of database query production. In Proceedings of the Human Factors Society (pp. 9-13). Santa Monica, CA: Human Factors Society.
- Harvard Medical School. (1996). FlyBase (World Wide Web URL http://cbbridges.harvard.edu:7081/). Cambridge, MA.
- Jackson Laboratory. (1996). Mouse Genome Database (World Wide Web URL http://www.informatics.jax.org/). Bar Harbor, ME.
- Johns Hopkins School of Medicine. (1996). Genome Database (GDB) (World Wide Web URL http://gdbwww.gdb.org/). Baltimore, MD.
- Landauer, T. K. (1995). The Trouble with Computers. Cambridge, MA: MIT Press.
- National Center for Genome Resources. (1996). Genome Sequence Database (GSDB) (World Wide Web URL http://www.ncgr.org/gsdb/). Santa Fe, NM.
- Netscape Communications Technical Document (1996). Persistent Client State - HTTP Cookies. (World Wide Web URL http://home.netscape.com/newsref/std/cookie_spec.html). Mountain View, CA.
- Newman, W.M., & Lamming, M.G. (1995). Interactive System Design. Wokingham, England: Addison-Wesley.
- Nielsen, J. (1993). Usability Engineering. Boston: Academic Press.
- Rice, J., Farquhar, A., Piernot, P.,& Gruber, T. (1996). Using the Web Instead of a Window System, In Proceedings of CHI'96 (pp. 103-110). Vancouver,B.C.: ACM.
- Schuler, D., & Namioka, A. (1993). Participatory Design: Principles and Practices. Hillsdale, NJ: Lawrence Erlbaum Assoc.
- University of Oregon Neuroscience Institute (1996). The FISH Net. (World Wide Web URL http://zfin.org). Eugene,OR.
- Yang, J. Y. & Kaiser, G. E. (1996). An Architecture for Intergrating OODBs with WWW, In Proceedings of Fitfth Intl. WWW Conference . Paris, France. (Also available at http://www5conf.inria.fr/fich_html/papers/P31/Overview.html.)
Notes
- This observation raises an obvious question: if Java supports the full expressive power of a modern interface programming environment, why not implement the entire interface as a Java applet, completely avoiding all constraints associated with HTML/HTTP? Several factors make this impractical. First, increased expressive power comes at the price of increased complexity. Implementing even relatively simple interfaces requires the sustained efforts of a skilled programmer; subsequent revisions compound this effort. We also found that the performance of Java applets varies widely, depending on the capabilities of client-side hardware and software. For these reasons, the ZFIN interface relies on Java only for those interface elements that can not be supported in any other way.
- Note that this enhancement is theoretically significant, upgrading the underlying model of the task space from a finite state automaton to a more powerful push-down automaton.
- Although one could conceivably implement functionally similar tools using only HTML (e.g. using clickmaps), the resulting interface would be extremely unwieldy, because every action requiring a response by the system would require a server transaction to refresh the display.
Screen Snapshots (Figures 4-8)