- Title
-
Developmental loss of NMDA receptors results in supernumerary forebrain neurons through delayed maturation of transit-amplifying neuroblasts
- Authors
- Napoli, A.J., Laderwager, S., Zoodsma, J.D., Biju, B., Mucollari, O., Schubel, S.K., Aprea, C., Sayed, A., Morgan, K., Napoli, A., Flanagan, S., Wollmuth, L.P., Sirotkin, H.I.
- Source
- Full text @ Sci. Rep.
|
|
|
|
|
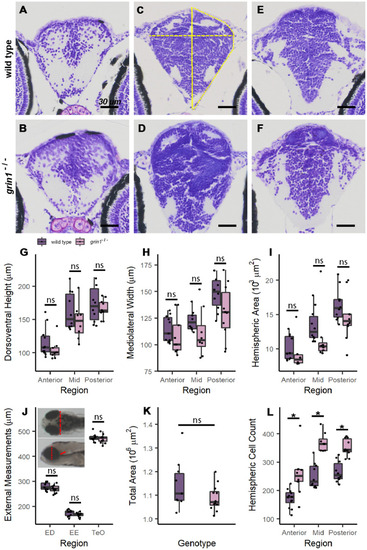
NMDA receptors suppress neuron proliferation in the forebrain. ( |
|
Radial glia are not increased in the forebrain of |
|
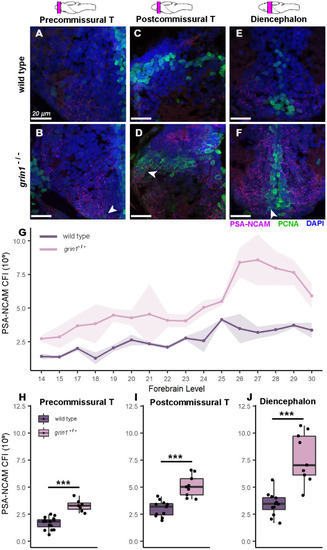
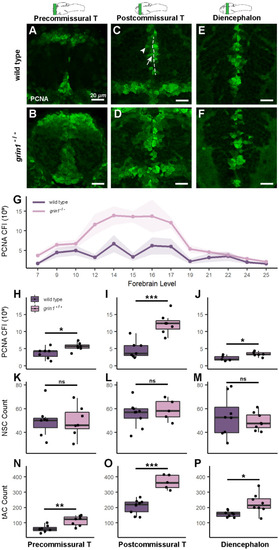
NMDA receptors suppress proliferation in neuroblasts (transit amplifying cells). ( |
|
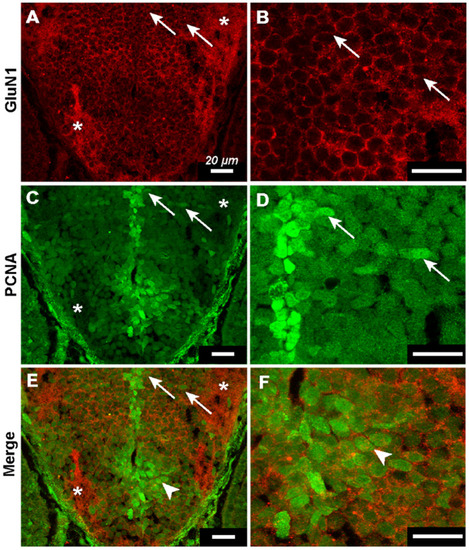
Transit amplifying cells express GluN1. ( |
|
KCC2 and PCNA expression are inversely correlated in the forebrain of wild-type fish. ( |
|
Loss of NMDA receptor function results in reduced KCC2 expression. ( |
|
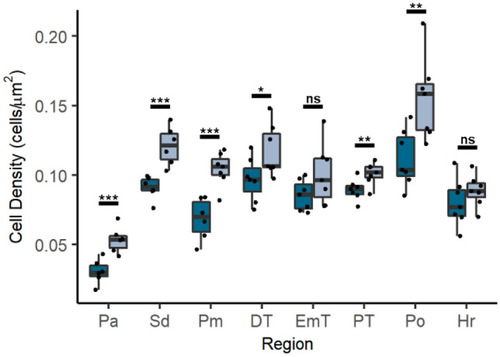
NMDA receptor ionotropic function is required suppression of neurogenesis. Box and whisker plot of cell density in 8 anatomical regions in control and MK-801 treated fish at 5 dpf. Significance |