- Title
-
Multi-focus averaging for multiple scattering suppression in optical coherence tomography
- Authors
- Zhu, L., Makita, S., Tamaoki, J., Lichtenegger, A., Lim, Y., Zhu, Y., Kobayashi, M., Yasuno, Y.
- Source
- Full text @ Biomed. Opt. Express
|
Schematics of trajectories of SS and MS photons with different focus depths; i.e., different defocus amounts. No matter the focal position, the SS photon is scattered only once and hence has the same path length. This results in the consistent phase of the SS signal after computational refocusing. Meanwhile, the trajectories of MS photons are scrambled by changes in the focal position. Hence, the phase of the MS signal is randomized even after the computational refocusing. |
|
Schematic (a) and photograph (b) of a scattering phantom. The phantom comprises glass slips A and D, a scattering layer B, which is a mixture of polystyrene micro-particles and ultrasound gel, and a glass plate C embedded in the scattering layer. The glass plate C provides a scattering-free area. A postmortem zebrafish at 30 days post fertilization (dpf) was used as a biological sample. The sample is shown in a color photograph (c) and a wide-field-of-view OCT intensity projection (d). The orange box around the belly region denotes the measurement area in the validation study of the MFA method. |
|
(a)–(c) and (d)–(f) show the B-scans and |
|
Cross sectional and |
|
(a) and (b) are B-scans at the same location of a phantom volume without and with refocusing, respectively. (c) and (d) show representative spatial auto-correlation functions of the linear-scaled images at a depth [dashed vertical lines in (e)] along both directions without and with applying refocusing, respectively, from which the FWHMs are measured. (e) shows the FWHMs along the depth, which are considered to be proportional to the speckle size. The first and second rows present the results without and with computational refocusing, respectively. Each column presents the results for a volume measured with different focal positions. The blue and red plots show the FWHMs for the fast-scanning (X) and slow-scanning (Y) directions. The B-scans (a) and (b) were taken from the volumes corresponding to the plots highlighted by green boxes. The used volumes are identical to those used for the MFA images in |
|
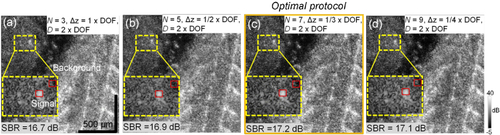
SBR plot against the number of averaged volumes |
|
|
|
Optical design schematics of the sample arm without (a) and with (b) the ETL. (c) Color photograph of the sample arm with the ETL. GS: galvanometric scanner, OBJ: objective, ETL: electrical tunable lens, and CM: collimator. Since the ETL is a refractive optical element and easy to align, switching between the two configurations without and with the ETL takes only a few minutes. (d) shows the simulation which reveals that the focus spot size becomes smaller as the focus is shifted away from the objective. The focus position is denoted by the distance from the bottom surface of the objective to the focal plane, the blue background denotes the focus shifting range |