- Title
-
Bone mineral density loci specific to the skull portray potential pleiotropic effects on craniosynostosis
- Authors
- Medina-Gomez, C., Mullin, B.H., Chesi, A., Prijatelj, V., Kemp, J.P., Shochat-Carvalho, C., Trajanoska, K., Wang, C., Joro, R., Evans, T.E., Schraut, K.E., Li-Gao, R., Ahluwalia, T.S., Zillikens, M.C., Zhu, K., Mook-Kanamori, D.O., Evans, D.S., Nethander, M., Knol, M.J., Thorleifsson, G., Prokic, I., Zemel, B., Broer, L., McGuigan, F.E., van Schoor, N.M., Reppe, S., Pawlak, M.A., Ralston, S.H., van der Velde, N., Lorentzon, M., Stefansson, K., Adams, H.H.H., Wilson, S.G., Ikram, M.A., Walsh, J.P., Lakka, T.A., Gautvik, K.M., Wilson, J.F., Orwoll, E.S., van Duijn, C.M., Bønnelykke, K., Uitterlinden, A.G., Styrkársdóttir, U., Akesson, K.E., Spector, T.D., Tobias, J.H., Ohlsson, C., Felix, J.F., Bisgaard, H., Grant, S.F.A., Richards, J.B., Evans, D.M., van der Eerden, B., van de Peppel, J., Ackert-Bicknell, C., Karasik, D., Kague, E., Rivadeneira, F.
- Source
- Full text @ Commun Biol
|
Regional plots for the four novel loci associated with SK-BMD ( Circles show GWAS meta-analysis P-values and position of SNPs for the overall meta-analysis ( |
|
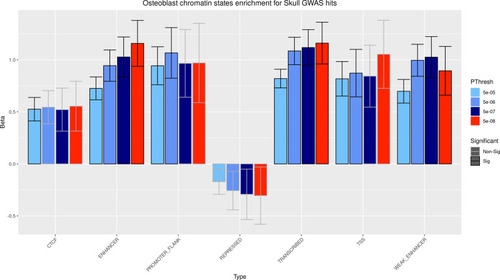
GARFIELD results for chromatin states enrichment analysis in osteoblasts. Enrichment significance was defined at |
|
DEPICT results for gene-set and cell/tissue enrichment analyses. |
|
Rapid functional evaluation of novel BMD associated genes in zebrafish identifies a role of novel gene |