- Title
-
Glucocorticoid effects in the regenerating fin reflect tissue homeostasis disturbances in zebrafish by affecting Wnt signaling
- Authors
- Fleischhauer, L., López-Delgado, A.C., Geurtzen, K., Knopf, F.
- Source
- Full text @ Front Endocrinol (Lausanne)
|
Quantification of fin regenerate length during the course of regeneration. Bars represent mean ± SD of fin outgrowth after amputation under different treatment conditions, measured at different times post amputation. Each dot represents one biological replicate. Parametric testing because of normal distribution of data. Statistical significance at 9 dpa, 14, dpa and 17 dpa was tested by two-tailed t-tests and at 21 dpa by |
|
Quantification of the |
|
Images of the mCherry-fluorescent Wnt-active zone in zebrafish fin regenerates. The |
|
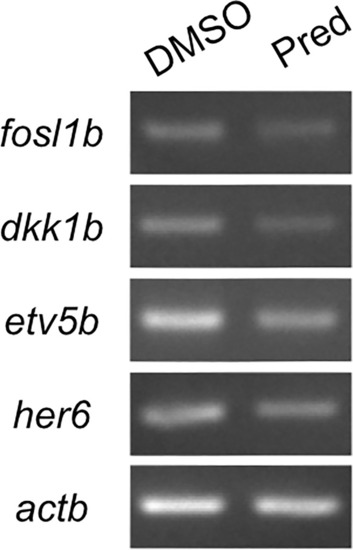
RT-PCR on Wnt, Fgf and Notch target genes and the Wnt signaling inhibitor |
|
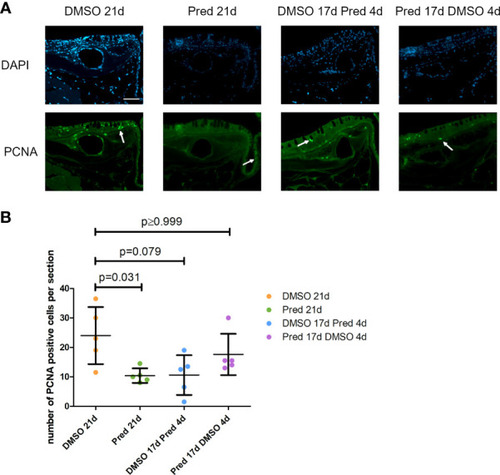
Quantification of PCNA positive cells along the skull bone of the zebrafish. |
|
Proliferation of scale forming cells after treatment. |
|
Cell proliferation in the telencephalon of zebrafish. |
|
Wnt signaling in the skin of zebrafish. |
|
Dkk1 expression in the skin of zebrafish. Immunohistochemical staining against Dkk1 and nuclear counterstain with DAPI in zebrafish that underwent 21 days of DMSO or 21 days of prednisolone treatment. 4 out of 5 zebrafish with prednisolone treatment showed stronger staining in the basal portion of skin than DMSO treated zebrafish (4 out of 5 DMSO treated zebrafish with weak staining in the basal portion of the skin). Scale bar 50 µm. Arrows point to brighter signal in the basal portion of the skin. Asterisks indicate mucous cells. n=5 (4 females, 1 male in DMSO 21d, 5 females in Pred 21d) in both groups with 5 sections per individual. |
|
Cell proliferation in the skin of zebrafish. |
|
Cell proliferation and goblet cell number in the crypts of the intestine. |
|
Absence of mCherry specific staining in Wnt-reporter |
|
Leukocytes in the zebrafish intestine. |