- Title
-
Acquisition of a large virulence plasmid (pINV) promoted temperature-dependent virulence and global dispersal of O96:H19 enteroinvasive Escherichia coli
- Authors
- Miles, S.L., Torraca, V., Dyson, Z.A., López-Jiménez, A.T., Foster-Nyarko, E., Lobato-Márquez, D., Jenkins, C., Holt, K.E., Mostowy, S.
- Source
- Full text @ MBio
|
Time-calibrated phylogeny of 92 Sequence Type (ST)99 genomes. BactDating was used to infer a time-calibrated phylogeny, incorporating the output from the recombination detection software, Gubbins. Blue diamonds indicate the internal nodes representing the most recent common ancestors (MRCA) of interest. Tip labels represent assembly barcodes correlating to the isolate accession in Enterobase. Tip labels in bold represent isolates that we tested |
|
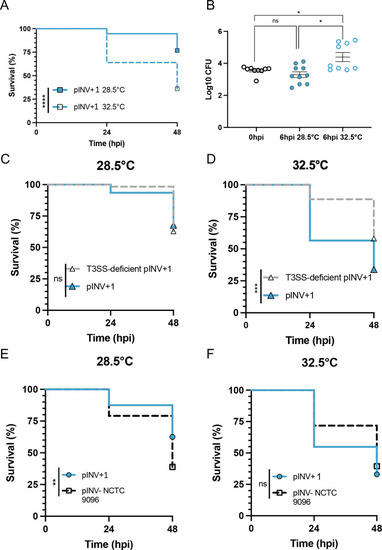
Temperature-dependent and -independent mechanisms of virulence in the ST99 group. Zebrafish larvae at 3 d post-fertilization were injected with 5,000 CFU of a representative pINV+ ST99 strain, a T3SS-deficient strain and an ancestral pINV− ST99 strain, before being separated for incubation at 28.5°C or 32.5°C. ( |