- Title
-
P1 Bacteriophage-Enabled Delivery of CRISPR-Cas9 Antimicrobial Activity Against Shigella flexneri
- Authors
- Huan, Y.W., Torraca, V., Brown, R., Fa-Arun, J., Miles, S.L., Oyarzún, D.A., Mostowy, S., Wang, B.
- Source
- Full text @ ACS Synth Biol
|
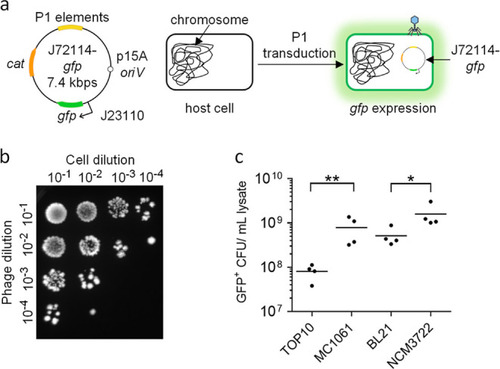
P1 J72114 phagemid as a delivery tool for transduction of a foreign genetic cassette into Gram-negative Enterobacteriaceae. (a) Schematic diagram of P1-J72114 phagemid, characterized by constitutive expression of gfp placed under the BBa_J23115 promoter. The phagemid contains the chloramphenicol acetyltransferase gene (cat), which confers a chloramphenicol-resistant phenotype to transduced or transformed cells. Transduced E. coli cells will retain the J72114-gfp phagemid, giving constitutive gfp expression. (b) Representative image showing the presence of GFP-positive E. coli NCM3722 cells after transduction with serially diluted phagemid lysates delivering the gfp expression cassette. Serial dilutions of recovered cells (101, 102, 103, and 104) were made and spotted onto LB agar supplemented with chloramphenicol. (c) Quantification of transducing units (phage particles containing phagemid sequences) in various lab strains of E. coli. Each data point represents a biological replicate and is the average of four technical repeats. Horizontal lines represent the group mean. The p-values were determined and adjusted by the Kruskal–Wallis test and Dunn’s multiple comparison tests, respectively, and significance was defined as p < 0.05. * represents p < 0.05, while ** represents p < 0.005, for comparisons between BL21 vs NCM3722 and TOP10 vs MC1061 and phagemid transductants, respectively. |
|
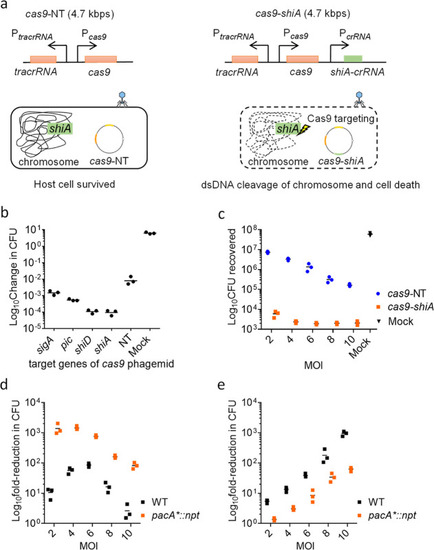
Spacer sequence-mediated lethality of E. coli MC1061::npt cells using npt-targeting cas9 phagemid. (a) Schematic diagrams showing the cas9 genetic construct, with or without npt-targeting crRNA (cas9-npt and cas9-NT, respectively) assembled onto the P1 J72114 phagemid. The presence of npt-targeting crRNA would target the Cas9 endonuclease chromosome of E. coli MC1061::npt cells, causing dsDNA cleavage of the chromosome and cell death. (b) Serial dilutions of transduced E. coli MC1061::npt were plated onto plain LB agar or LB agar supplemented with kanamycin. Data were plotted as change(s) in CFU as compared to input CFU (approximately 107 cells per reaction) used for infection. (c) Quantification of chloramphenicol-resistant CFUs recovered, after treatment with cas9-NT or cas9-npt phagemid lysates. Each data point represents a biological replicate and is the average of four technical repeats. A MOI of 5 P1 transducing units per bacterial cell was used for all infections. Mock infections involved treating E. coli cells with SM buffer. Horizontal bars represent the group mean. The p-values (between non-targeting and targeting phagemid treatments) were determined using a Kruskal–Wallis test, with the significance defined by p < 0.05. p < 0.0005 between targeting phagemid and nontargeting phagemid treatments is shown as ***. |
|
Genetic modification of the pac site, within the pacA coding sequence of the wildtype P1 bacteriophage genome to reduce wildtype P1 phage DNA packaging. (a) Schematic diagram showing the minimal pac sequence of the P1 phage genome, situated within the pacA coding sequence. Methylation sites, consisting of hexameric repeats, HEX4 and HEX3, are shown in black, the IHF binding site is shown in green, and the pac cleavage site is shown in orange. Synonymous mutations were introduced onto the hexameric repeats to disrupt the binding of PacA and PacB. A Comparison with the wildtype pac DNA sequence is shown, with mutated DNA bases indicated in red. (b) Schematic diagram showing the processing of the pac site, involving the binding of PacA and PacB to the hexameric repeats, as well as IHF and HU binding, which was proposed to give a bent structure of the pac site. Pacase (both PacA and PacB) enzymatic activity then leads to cleavage of the pac site.34,35 Synonymous mutations introduced onto the hexameric repeats of the pac site lead to a reduction in pacase binding, reducing the processing and packaging of the P1 mutant pacA*::npt P1 DNA. (c) Quantification of wildtype P1 phage (in blue) and P1 transducing units (in orange) titers of lysates prepared from wildtype (WT) and pacA*::npt EMG16 cells harboring J72114 cas9-NT phagemid without spacer sequence(s) targeting E. coli or S. flexneri chromosomal sequences. (d) Ratio of wildtype P1 phage to P1 transducing units was calculated and plotted. Each data point represents a biological replicate and is the average of four technical repeats. Horizontal bars represent the group mean. The p-values (between wildtype and pacA*::npt lysates) were determined using a two-tailed unpaired t-test with significance defined by p < 0.05. p < 0.0005 between the P1 phage titer of lysates prepared from wildtype and pacA* EMG16 lysogen, as well as the PFU/TU ratio between lysates prepared from wildtype and pacA* EMG16 lysogen are shown as ***. |
|
Cas9-mediated lethality of S. flexneri using chromosomal-targeting J72114 cas9-shiA phagemid. (a) Schematic diagram showing the cas9 genetic construct of P1 J72114 phagemid, with spacer sequence targeting chromosomal gene(s) (i.e., shiA) of S. flexneri (in green). Upon transduction, the presence of crRNA with a spacer sequence complementary to the chromosomal genes of S. flexneri (ct-crRNA) would cause dsDNA cleavage of the chromosome via Cas9 endonuclease, leading to cell death. (b) J72114 cas9 phagemid with spacer sequence(s) targeting the sigA, pic, shiD, and shiA chromosomal genes of S. flexneri 2a 2457O. Lysates were prepared from the wildtype EMG16 cell line. Crude lysates were used for transduction of S. flexneri 2A 2457O, with a MOI of 10 wildtype P1 phages (equivalent to 5 transducing units) to 1 bacterial cell. Data were plotted as change(s) in CFU as compared to input CFU (approximately 1 × 107 cells per reaction) used for infection. (c) Spacer sequence-mediated lethality of S. flexneri 5a MT905 cells with chromosomal-targeting cas9-shiA (in orange) and nontargeting cas9-NT phagemids (in blue) in lysates prepared from the pacA*::npt EMG16 cell line. MOIs of 2.0, 4.0, 6.0, 8.0, and 10.0 were used. Data were plotted as changes in CFU as compared to input CFU (approximately 1 × 107 cells per reaction) used for infection. The number of CFU recovered from the mock infection was shown in black. (d) Cas9 spacer sequence-mediated lethality effect of cas9-shiA phagemid and (e) Nonspacer sequence-mediated lethality effect of cas9 phagemid lysate prepared from wildtype (WT, in black) and pacA*::npt EMG (in orange) cell lines. MOIs of 2, 4, 6, 8, and 10 P1 transducing units to 1 bacterial cell for pacA*::npt EMG lysates were used. For wildtype EMG16 lysates, MOIs of 2.0, 4.0, 6.0 , 8.0, and 10.0 wildtype P1 phages per bacterial cell were used, and the wildtype P1 phage to P1 transducing units is approximately 2 for lysates prepared from the wildtype EMG16 cell line. The cas9-shiA-mediated lethality effect was quantified by measuring the reduction in CFU recovered after treatment with cas9-shiA and cas9-NT phagemids. Mock infections involved treating S. flexneri cells with SM buffer. The reduction in CFU recovered between cas9-NT phagemid treatment and the input cells used for infection (approximately 1 × 107 cells per reaction) would show the nonspacer sequence-mediated lethality effect of P1 phage lysates at all four MOIs tested. Each data point represents a biological repeat and is the average of four technical repeats. Horizontal bars represent the group mean. |
|
Spacer sequence-mediated lethality of Shigella flexneri 5a M90T in vivo using chromosomal-targeting cas9-shiA phagemid. (a) Enumeration of S. flexneri 5a M90T CFU at 6 h post-infection. n = 9 (targeting); 9 (empty) larvae (cumulated from three independent experiments). (b) Measurement of the survival rate of infected zebrafish larvae at 24, 48, and 72 h post-infection (hpi), treated with cas9-shiA phagemid (orange) or with the nontargeting cas9-NT phagemid (blue). n = 71 (targeting); 74 (empty) larvae (cumulated from three independent experiments). Differences in bacterial load were tested using an unpaired t-test on log 10-transformed data, while differences in survival were tested using a log-rank (Mantel–Cox) test. p < 0.05 is considered statistically significant. p < 0.005 between targeting phagemid and nontargeting phagemid treatments are shown as **. (c) Schematic diagram showing the reduction of S. flexneri infection bacterial load via the spacer sequence-mediated lethality effect of cas9-shiA phagemid. The reduced bacterial load would promote the survival of infected larvae. |