- Title
-
Comparative Study of the Molecular Characterization, Evolution, and Structure Modeling of Digestive Lipase Genes Reveals the Different Evolutionary Selection Between Mammals and Fishes
- Authors
- Tang, S.L., Liang, X.F., He, S., Li, L., Alam, M.S., Wu, J.
- Source
- Full text @ Front Genet
|
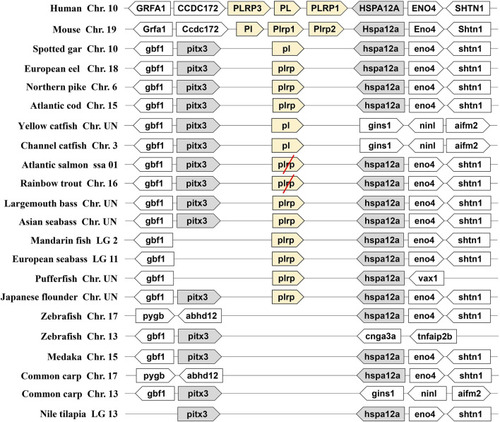
Synteny of pancreatic lipase genes (light yellow) in mammals and fishes. The conserved neighborhood genes ( |
|
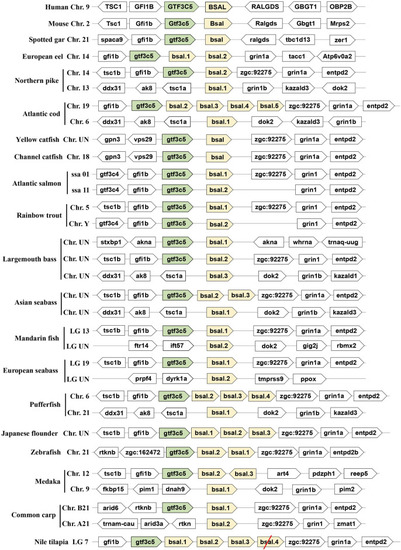
Synteny of bile salt-activated lipase genes (light yellow) in mammals and fishes. The conserved neighborhood genes ( |
|
Phylogenetic relationship of pancreatic lipase genes in mammals and fishes with based on the ML method. The bootstrap test (1000 replicates) scores are shown with numbers. The blackline represents the group of |
|
Phylogenetic relationship of bile salt-activated lipase genes in mammals and fishes with based on the ML method. The bootstrap test (1000 replicates) scores are shown with numbers. The black line represents the group of |
|
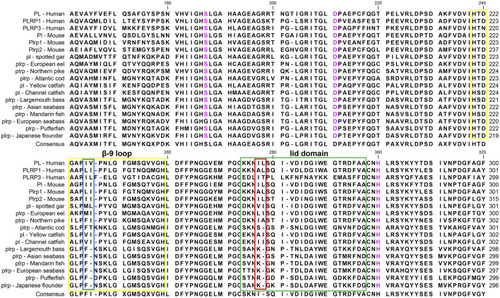
Partial amino acid sequence alignments of pancreatic lipases for humans and fishes. Boxes in green and yellow represent the lid domain and β-9 loop of pancreatic lipase, respectively. Active site triad residues Ser, Asp, and His were marked with pink. The red and blue boxes represent the absence of Ile and Leu in the lid domain and β-9 loop, respectively. The dashes (-) represent gaps found upon sequence alignment. Numbers refer to the amino acid were located at the end of each line. |
|
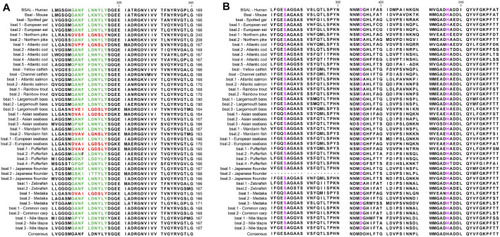
Partial Amino acid sequence alignments of bile salt-activated lipases for humans and fishes. |
|
3D structures of pancreatic lipase in human, mouse, and partial fish species. The lid domain and β-9 loop of pancreatic lipase were marked with green and yellow, respectively. The regions of Ile and Leu (or replaced by other amino acids) in the lid domain and β-9 loop were marked with red and blue, respectively. Active site triad residues Ser, Asp, and His were marked with pink. |
|
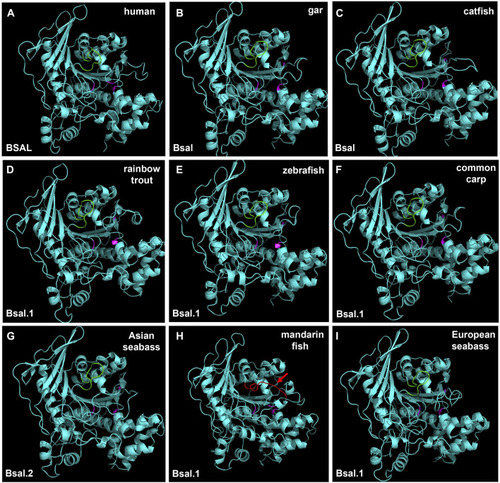
3D structures of bile salt-activated lipases in human and partial fish species. The region of amino acid residues in the bile salt-binding site (GANFLXNYLY) was marked with green. Active site triad residues Ser, Asp, and His were marked with pink. The red arrow represents the no loop structure that existed in mandarin fish |
|
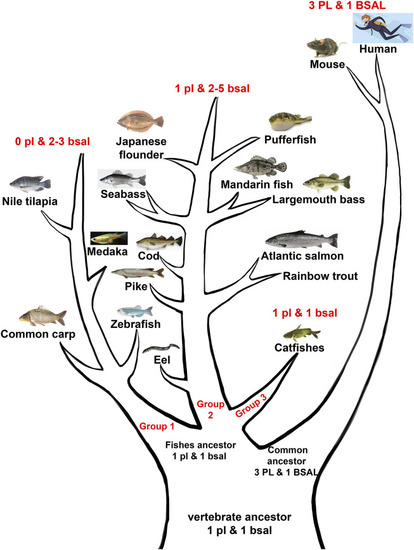
Abridged general view of digestive lipases during the evolution among mammals and fishes. The vertebrate ancestor might have one |