- Title
-
Elucidation of the genetic causes of bicuspid aortic valve disease
- Authors
- Gehlen, J., Stundl, A., Debiec, R., Fontana, F., Krane, M., Sharipova, D., Nelson, C.P., Al-Kassou, B., Giel, A.S., Sinning, J.M., Bruenger, C.M., Zelck, C.F., Koebbe, L.L., Braund, P.S., Webb, T.R., Hetherington, S., Ensminger, S., Fujita, B., Mohamed, S.A., Shrestha, M., Krueger, H., Siepe, M., Kari, F.A., Nordbeck, P., Buravezky, L., Kelm, M., Veulemans, V., Adam, M., Baldus, S., Laugwitz, K.L., Haas, Y., Karck, M., Mehlhorn, U., Conzelmann, L.O., Breitenbach, I., Lebherz, C., Urbanski, P., Kim, W.K., Kandels, J., Ellinghaus, D., Nowak-Goettl, U., Hoffmann, P., Wirth, F., Doppler, S., Lahm, H., Dreßen, M., von Scheidt, M., Knoll, K., Kessler, T., Hengstenberg, C., Schunkert, H., Nickenig, G., Nöthen, M.M., Bolger, A.P., Abdelilah-Seyfried, S., Samani, N.J., Erdmann, J., Trenkwalder, T., Schumacher, J.
- Source
- Full text @ Cardiovasc. Res.
|
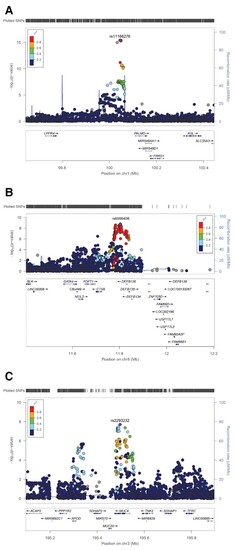
Regional association plots of BAV risk loci on chromosome 1p21 near PALMD (A), 8p23 near GATA4 (B) and 3q29 within MUC4 (C). At each locus, associations [−log10(P-values)] are shown for SNPs flanking 400 kb on either side of the lead associated SNP. The lead variant is indicated by the corresponding rs-number. Other markers at each locus are displayed by different colours, which indicate different levels of LD (r2) to the lead SNP. Furthermore, all annotated genes within each region are shown with arrows indicating their transcription direction. |
|
Zebrafish muc4 knockout using CRISPR/Cas9. (A–D) Maximum intensity projections from confocal z-stack images of hearts at 58 hpf. (A’–D’) Single confocal z-section images of the AVC region. Endocardial cells in luminal positions (arrowheads) at the AVC are marked by kdrl:GFP and Alcam.45 Endocardial cells that have already invaded the extracellular matrix and are in abluminal positions are marked with asterisks. (A’) While in WT control embryos many endocardial cells are in abluminal positions, some of the 15 muc4 crispants with a complete truncation of the genomic locus exhibit a delayed ingression of endocardial cells into the cardiac jelly. The severity of the phenotypes varied according to the following classes: (B’) Class 1 embryos (3 of 15) lack an endocardial cell ingression by that stage; (C’) Class 2 embryos (4 of 15), have only a single endocardial cell ingressing into the cardiac jelly at that stage; (D’) in muc4 crispants of class 3 (8 of 15), a few endocardial cells have ingressed into the cardiac jelly. V, ventricle; A, atrium; AVC, atrioventricular canal. Scale bars are 20 µm (A–D) and 10 µm (A’–D’). (E) Schematic representation of the zebrafish muc4 locus and gRNA binding sites (red arrows). Black arrows indicate the positions of primers used to assess the efficacy of CRISPS-induced knockout by PCR. (E’) Single embryo PCR products from control embryos (ctrl) and embryos injected with a mixture of four gRNAs and Cas9 protein (muc4_crispants). Red boxes are indicating samples that lack any WT genomic amplification bands. Among 58 injected embryos that were genotyped, 15 had a large truncation of the genomic region and did not contain any WT band. Only these crispants were used for phenotypic characterization. MW, molecular weight. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |