- Title
-
HIVEP1 Is a Negative Regulator of NF-κB That Inhibits Systemic Inflammation in Sepsis
- Authors
- Matsumoto, H., Scicluna, B.P., Jim, K.K., Falahi, F., Qin, W., Gürkan, B., Malmström, E., Meijer, M.T., Butler, J.M., Khan, H.N., Takagi, T., Ishii, S., Schultz, M.J., van de Beek, D., de Vos, A.F., van 't Veer, C., van der Poll, T.
- Source
- Full text @ Front Immunol
|
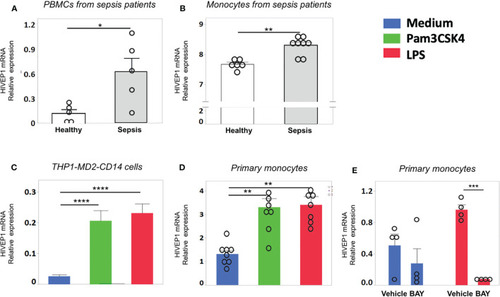
HIVEP1 mRNA expression is increased in patients with sepsis and upon stimulation with bacterial agonists |
|
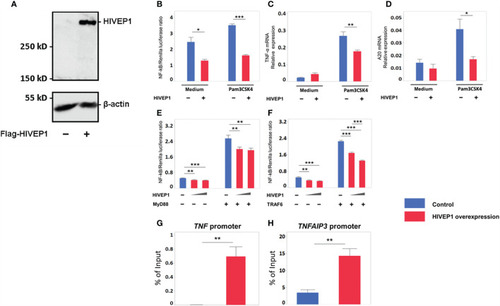
Overexpression of HIVEP1 inhibits NF-κB activity upon Toll-like receptor stimulation and binds to the TNF and TNFAIP3 promoter regions. |
|
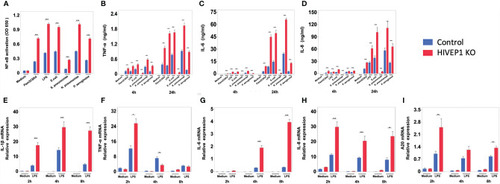
HIVEP1 deficient human monocytic cells show increased NFκB activity and proinflammatory cytokine production upon stimulation with TLR ligands or bacteria. HIVEP1 deficient and control human monocytic THP1-MD2-CD14 cells were stimulated with PAM3CSK4 (1 µg/mL), LPS (100 ng/mL), heat-killed |
|
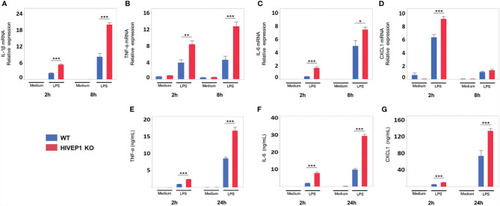
HIVEP1-deficient murine macrophages show increased proinflammatory cytokine production upon stimulation with LPS. Bone marrow derived macrophages from |
|
RNA-sequencing of HIVEP1 deficient THP-1 cells before and after LPS stimulation. |
|
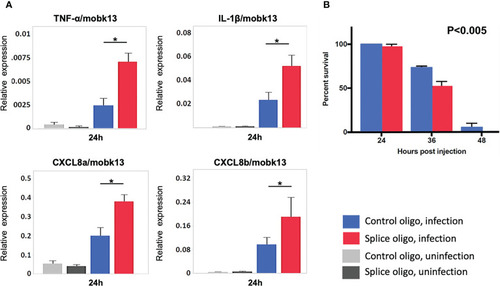
HIVEP1 knockdown increases systemic inflammation and impairs survival in zebrafish embryos with sepsis. |