- Title
-
Growth Hormone Overexpression Induces Hyperphagia and Intestinal Morphophysiological Adaptations to Improve Nutrient Uptake in Zebrafish
- Authors
- Meirelles, M.G., Nornberg, B.F., da Silveira, T.L.R., Kütter, M.T., Castro, C.G., Ramirez, J.R.B., Pedrosa, V., Romano, L.A., Marins, L.F.
- Source
- Full text @ Front. Physiol.
|
|
|
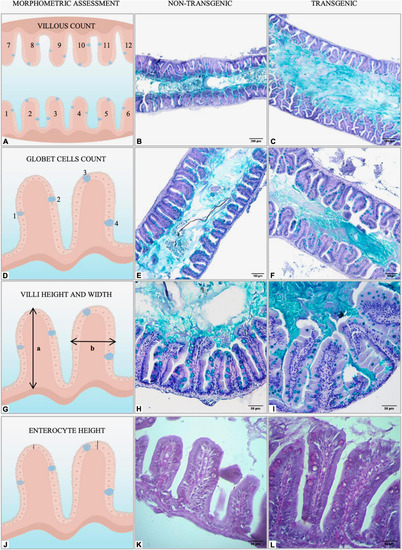
Graphical representation of morphometric analysis performed in the intestine of non-transgenic and |
|
Growth performance and feed intake of |
|
Intestinal absorptive area of |
|
Ultrastructure of the intestinal microvilli of |
|
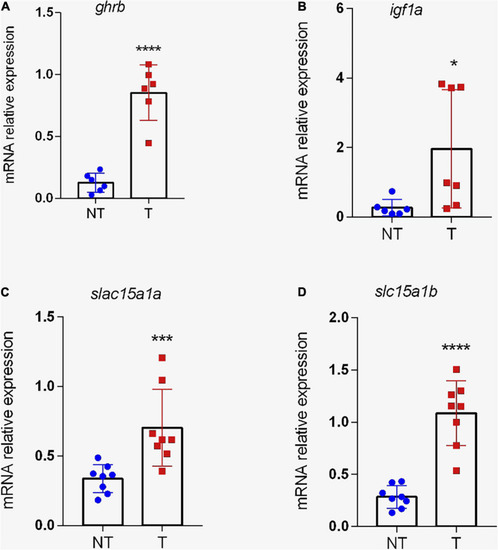
Expression of genes related to growth and peptide transport in the intestine of transgenic zebrafish ( |