- Title
-
FGF-2 promotes angiogenesis through a SRSF1/SRSF3/SRPK1-dependent axis that controls VEGFR1 splicing in endothelial cells
- Authors
- Jia, T., Jacquet, T., Dalonneau, F., Coudert, P., Vaganay, E., Exbrayat-Héritier, C., Vollaire, J., Josserand, V., Ruggiero, F., Coll, J.L., Eymin, B.
- Source
- Full text @ BMC Biol.
|
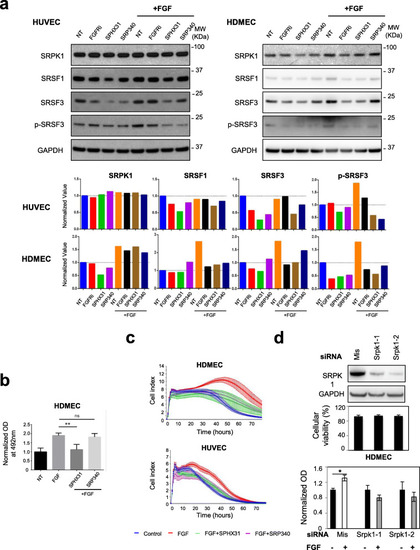
FGF-2 increases SRSF1, SRSF3 and SRPK1 protein levels in primary endothelial cells. |
|
FGF-2-mediated HUVEC survival requires SR proteins accumulation and phosphorylation. |
|
FGF2-mediated endothelial cells proliferation and survival requires SRPK1 activity. |
|
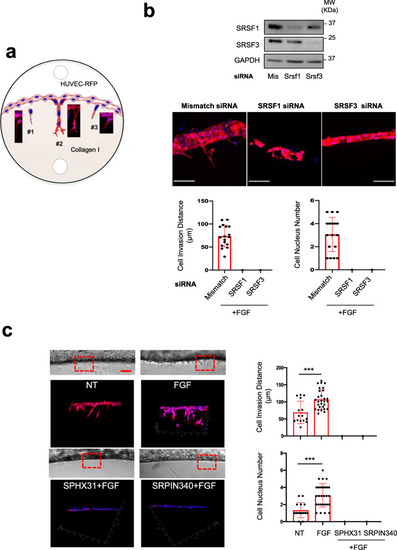
SRSF1/SRSF3 knockdown and SRPK1 inhibitors abrogate HUVEC-RFP invasion and sprouting in 3D collagen matrix. |
|
Endothelial cells accumulate SRSF1/3 and SRPK1 proteins upon FGF-2 treatment in an in vivo sponge assay. |
|
SRPIN340 treatment perturbs intersegmental vessels (ISV) sprouting and DLAV formation in zebrafish embryos. |
|
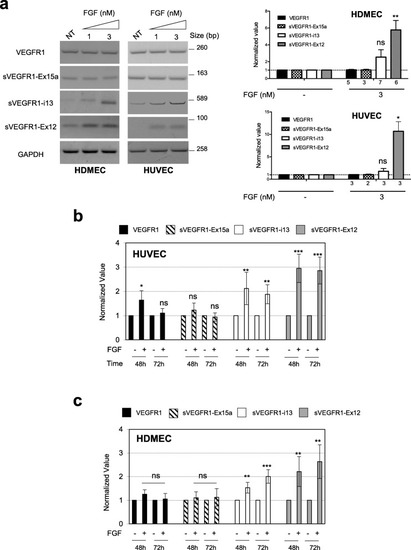
FGF-2 leads to the accumulation of sVEGFR1 splice variants in endothelial cells. |
|
FGF-2 mediates accumulation of sVEGFR1 splice variants through a SRSF1/SRSF3/SRPK1-dependent axis. a HDMEC were transfected with control siRNA (mis) or specific SRSF1 (Srsf1) or SRSF3 (Srsf3) siRNA and treated with 3nM FGF-2 for 72 h. |
|
sVEGFR1-ex12 contributes to FGF-2 pro-angiogenic functions in endothelial cells. |
|
sVEGFR1-ex12 is a poor prognosis marker in squamous lung carcinoma patients. |
|
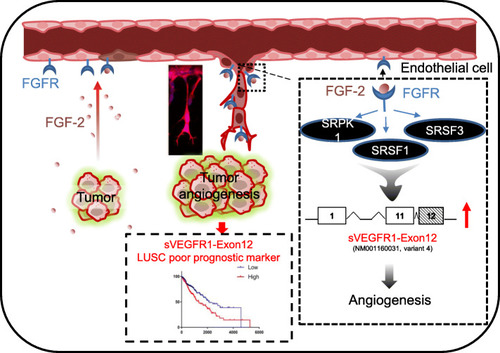
Graphical abstract for FGF-2-dependent regulation of sVEGFR1 splice variants in endothelial cells and its contribution to angiogenesis and lung tumorigenesis. In endothelial cells, FGF-2 stimulates a SRPK1/SRSF1/SRSF3 signaling pathway that controls VEGFR1 splicing in favor of sVEGFR1 splice variants, in particular sVEGFR1-ex12 (variant 4), that contribute to FGF-2 pro-angiogenic functions. In squamous lung carcinoma patients (LUSC), elevated sVEGFR1-ex12 usage value correlates with FGF-2/FGFR1 mRNA levels and with poor prognosis, thereby supporting a role of the FGF-2/FGFR1/sVEGFR1-ex12 signaling network in both physiological and pathological angiogenesis |