- Title
-
Metabolomic and transcriptomic profiling of adult mice and larval zebrafish leptin mutants reveal a common pattern of changes in metabolites and signaling pathways
- Authors
- Ding, Y., Haks, M.C., Forn-Cuní, G., He, J., Nowik, N., Harms, A.C., Hankemeier, T., Eeza, M.N.H., Matysik, J., Alia, A., Spaink, H.P.
- Source
- Full text @ Cell Biosci.
|
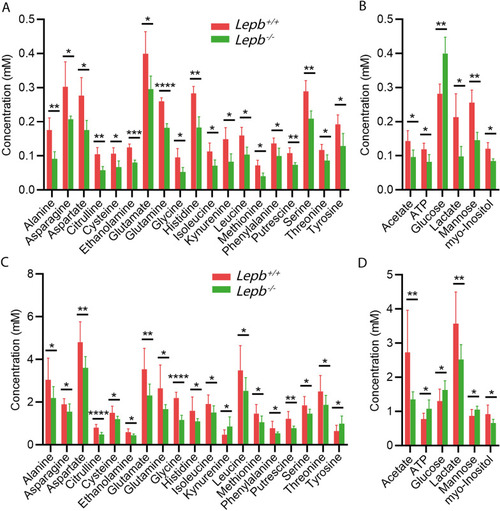
Metabolic profiles of blood from |
|
One-dimensional 1H NMR spectra and PLS-DA analysis of extracted |
|
One-dimensional 1H HR-MAS NMR spectra and PLS-DA analysis of intact |
|
Quantification of the common 25 metabolites that are significantly changed in zebrafish larvae. |
|
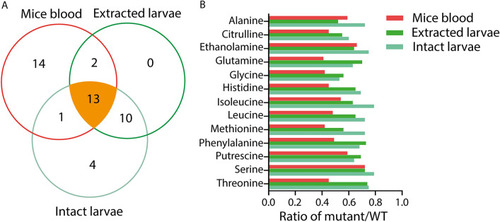
Common biomarkers for |
|
Lipid profiles of |
|
Transcriptome signature sets of mice and zebrafish larvae. A A Volcano plot showing a graphical representation of the significance (p < 0.05) in ob/ob mice head compared to C57BL/6 mice head. The transcripts with fold change over 1.5 are highlighted in red. Fifteen significant genes in mice head out of the fold change in X axis are excluded to make the graph look well. B A Venn diagram showing the comparison of the number of significantly changed genes between ob/ob mice head and mice liver published by Kokaji et al. C The top eight GO terms of biological process (BP) with lowest p adjusted values and highest numbers of genes representatives in mice head and the overlap of B. GO gene ontology. D Number of genes in classification of GO term proteolysis in the signature set of mice head. E A Volcano plot showing a graphical representation of the significance (p < 0.05) in lepb mutant zebrafish larvae compared to wild type siblings. The transcripts with fold change over 1.5 are highlighted in green. Twenty-two significant genes in zebrafish larvae out of the fold change in X axis are excluded to make the graph look well. F A Venn diagram showing the comparison of the number of significantly changed genes from human homologs of the signature gene sets of zebrafish larvae and ob/ob mice head. G The top eight GO terms of BP with lowest p adjusted values and highest numbers of genes representatives in zebrafish larvae and the overlap of F. H Number of genes in classification of GO term proteolysis in the signature set of zebrafish larvae |
|
Genes involved in arachidonic acid pathway in human orthologs of the three transcriptome signature sets. Dashed lines means indirect regulation. Red color represents genes significantly changed in |