- Title
-
An atlas of seven zebrafish hox cluster mutants provides insights into sub/neofunctionalization of vertebrate Hox clusters
- Authors
- Yamada, K., Maeno, A., Araki, S., Kikuchi, M., Suzuki, M., Ishizaka, M., Satoh, K., Akama, K., Kawabe, Y., Suzuki, K., Kobayashi, D., Hamano, N., Kawamura, A.
- Source
- Full text @ Development
|
Zebrafishhoxclusters and isolation of hoxaa cluster-deleted mutants. (A) The zebrafish possesses 48 hox genes in seven hox clusters. (B) Strategy for deletion of the genomic region corresponding to the hox cluster. Two dgRNAs (arrowheads), which recognize hox genes located at both ends of hox clusters, were used to delete the target genomic region. (C) Schematic of the hoxaa cluster locus. Six hox genes in the hoxaa cluster are depicted as white rectangles. The arrow shows the orientation of the transcription of each hox gene. Two black arrowheads indicate the genomic loci of the target sequence of dgRNAs, which were used to delete the genomic region of the hoxaa cluster. (D) Large genomic deletion in the hoxaa cluster mutant. The flanking sequences of two dgRNAs-target are shown. The target sequence of hoxaa-5′ dgRNA is exon 1 of hoxa13a (blue letters), and hoxaa-3′ dgRNA is targeted to exon 1 of hoxa1a (green letters). The hoxaa mutant possessing a ∼56.4 kb deletion and a 27 bp insertion (black letters) was isolated. This indel mutation resulted in frameshift of hoxa13a. (E) PCR-based genotyping of embryos obtained by intercross between hoxaa+/− fish. Genotyping was carried out with the three primers shown by blue arrowheads in C. The sequences of primers and PCR cycles are shown in Table S3. (F) Specific deletion of the hoxaa cluster was confirmed by PCR. The genomic DNA extracted from wild-type and hoxaa−/− embryos was used as a template for PCR. The sequences of the primers used in this analysis are listed in Table S4. |
|
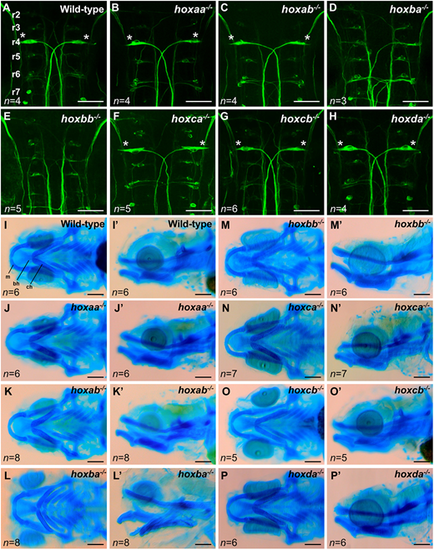
Reticulospinal neurons and craniofacial cartilage in zebrafishhoxcluster mutants. (A-H) Reticulospinal neurons in the hindbrain at 48 hpf were visualized by whole-mount immunostaining using anti-neurofilament RMO-44 antibody. Dorsal view of flat-mounted specimens after removal of the yolk. Mauthner cells located at r4 are indicated by asterisks. Hemizygous mutants for each hox cluster appeared indistinguishable from the wild-type. (I-P′) Craniofacial cartilages of zebrafish larvae at 5 dpf were stained with Alcian Blue. I-P show ventral view and I′-P′ show lateral view. bh, basihyal; ch, ceratohyal; m, Meckel's cartilage. Scale bars, 50 µm (A-H); 100 µm (I-P). PHENOTYPE:
|
|
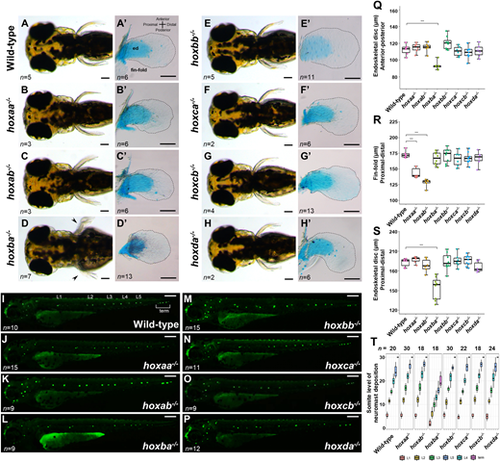
Pectoral fins and lateral line neuromasts in zebrafishhoxcluster mutants. (A-H) Pectoral fins of live hox cluster mutants at 3 dpf. Dorsal view. Arrowheads in D indicate pectoral fin malformation in hoxba cluster mutants. (A′-H′) Dissected pectoral fins at 5 dpf. Alcian Blue staining was performed to show cartilage cells (endoskeletal disc; ed). Hemizygous mutants for each hox cluster appeared indistinguishable from the wild-type. The margin of the fin-fold is emphasized by a dashed line. (I-P) Neuromasts in lateral lines were visualized in hox cluster mutants at 3 dpf. Posterior lateral line neuromasts are indicated from anterior-to-posterior as L1, L2, L3, L4, L5, and terminal (term) neuromasts. Hemizygous mutants for each hox cluster appeared indistinguishable from the wild-type. (Q) Comparison of the lengths of the endoskeletal disc along the anterior-posterior axis at the center of the fin. (R) Comparison of fin-fold lengths along the proximodistal axis in hox cluster mutants. (S) Comparison of the lengths of the endoskeletal disc along the proximodistal axis at the center of the fin. Box plot percentiles are: line, median; box, 25th and 75th percentiles; whiskers, 0th-25th and 75th-100th percentiles. The error bar represents the standard error. ***P<0.001 (two-tailed Student's t-test). (T) Positions of posterior lateral line neuromasts, which are located relative to the somite number, are compared in hox cluster mutants. Neuromasts located on both sides of the larvae were examined. Scale bars: 50 µm (A-H,I-P); 100 µm (A′-H′). PHENOTYPE:
|
|
Anterior homeotic transformation of the Weberian apparatus and pleural-to-caudal vertebrae inhoxcluster mutants. (A-R) The anteriormost vertebrae including the Weberian apparatus in wild-type zebrafish and hox cluster mutants were examined by micro-CT scanning and are shown as 3D-VR images. The Weberian apparatus is composed of the four ossicles including the tripus (tri), intercalarium (ic), scaphium (sc) and claustrum (cl), in the anteriormost vertebrae. Other ossicles such as the os suspensorium (os), transverse process of vertebra 4 (tp4) and lateral process (lp) are indicated in the panels. (S-X) Pleural ribs in wild-type and hox cluster mutants (n=7 for wild-type; n=4 for hoxaa−/−; n=2 for hoxbb−/−; n=3 for hoxca−/−; n=4 for hoxcb−/−; n=2 for hoxda−/−) were subjected to micro-CT scan analysis, and representative images are shown. The number represents the position of a vertebra from the first vertebra. The yellow number indicates the vertebra possessing the pleural ribs. The white number indicates the vertebra possessing the short ribs. The red number indicates the vertebra possessing the hemal arch that extends to the anteriormost radial in the anal-fin ray. In Table S1, the vertebral phenotypes are summarized and two hoxca−/− fish stained with Alizarin Red were included. hoxbb−/− and hoxda−/− are male and wild-type, hoxaa−/−, hoxca−/− and hoxcb−/− are female. Movies 1-10 show micro-CT scans. Scale bars: 1 mm (A-R); 2 mm (S-X). PHENOTYPE:
|
|
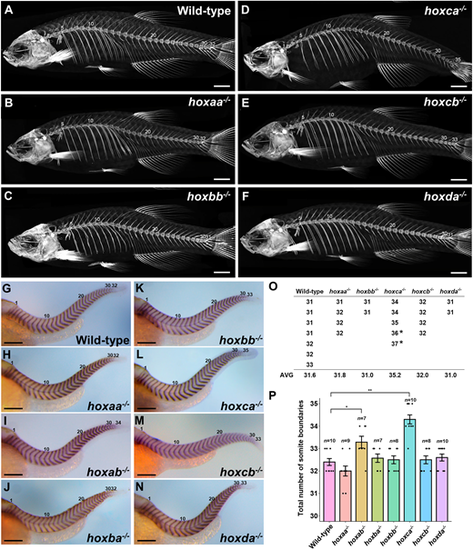
Total numbers of vertebrae and somite boundaries inhoxcluster mutants. (A-F) Whole-body skeletons were analyzed by micro-CT scanning. Adult fish (n=7 for wild-type; n=4 for hoxaa−/−; n=2 for hoxbb−/−; n=3 for hoxca−/−; n=4 for hoxcb−/−; n=2 for hoxda−/−) were examined by micro-CT scanning, and representative images are shown. (G-N) Expression patterns of the segment boundary marker, xirp2a/cb1045, in hox cluster homozygous embryos at 2 dpf. Lateral views. The number of somite boundaries from the anteriormost boundary is indicated. After staining, genotyping was performed. Hemizygous mutants for each hox cluster were indistinguishable from the wild-type. (O) Total numbers of vertebrae in hox cluster mutants. The numbers of vertebrates were counted using micro-CT scan images. In hoxca cluster mutants, two adult fish stained with Alizarin Red were included (indicated by the asterisk). (P) Comparison of the total numbers of somite boundaries in zebrafish hox cluster mutants. The numbers of somite boundaries in the mutants were compared by counting the segment boundaries stained with xirp2a/cb1045. Error bars represent the standard error. *P<0.05, **P<0.01 (two-tailed Student's t-test). Scale bars: 2 mm (A-F); 200 µm (G-N). EXPRESSION / LABELING:
PHENOTYPE:
|
|
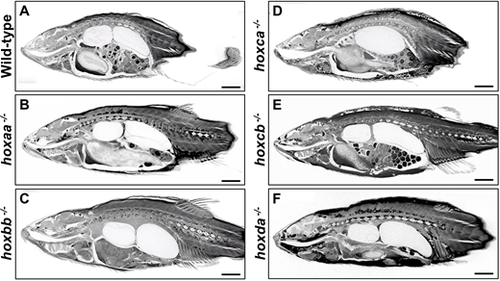
Whole-body tissues ofhoxcluster mutants. (A-F) Whole-body tissues were analyzed by micro-CT scanning. After the micro-CT scanning for skeleton analysis, soft tissues of the same specimens were stained with Lugol's solution, and the stained tissues were subject to micro-CT scanning. Adult males (hoxbb−/− and hoxda−/−) and females (wild-type, hoxaa−/−, hoxca−/− and hoxcb−/−) were used. Eggs are seen in the abdomen of female fish. Movies 11-20 show micro-CT scan 3D movies of transverse and sagittal sections. Scale bars: 2 mm. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|