- Title
-
Dynamic Polarization of Rab11a Modulates Crb2a Localization and Impacts Signaling to Regulate Retinal Neurogenesis
- Authors
- Clark, B.S., Miesfeld, J.B., Flinn, M.A., Collery, R.F., Link, B.A.
- Source
- Full text @ Front Cell Dev Biol
|
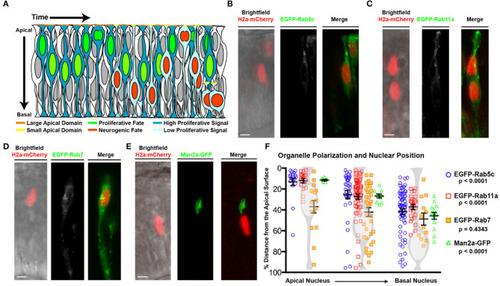
Organelle positioning during interkinetic nuclear migration. (A) Schematic of cellular features correlated with neurogenic and proliferative RPCs, including nuclear position, apical domain size, and proliferative signaling. (B–E) Examples of genetic mosaics of transplanted cells with H2a-mCherry labeled nuclei and endocytic organelles marked by EGFP-fusion proteins. (B) Early endosome (EGFP-Rab5c) localization in cells with apical nuclei. (C) Recycling endosome (EGFP-Rab11a) localization. (D) Late endosome (EGFP-Rab7) localization, and (E) localization of the medial Golgi apparatus (Man2a-GFP). (F) Quantification of the distance of organelles from the apical surface when nuclei are positioned apically (<25% of apical-basal distance), middle (25–50% of apical-basal distance), or basally (>50% of apical-basal distance). Data represent individual organelle positioning with mean and SEM indicated for each organelle type for each bin of nuclear positions from >10 cells/nuclear position bin from >5 embryos/genotype. Statistics represent results of a One-way ANOVA. Scale bars in (B–E) represent 10 μm. |
|
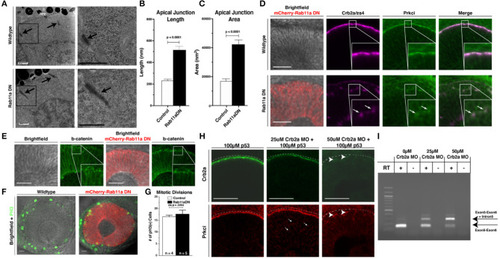
Rab11aDN expression causes Crb2a and Prkci mis-localization, but maintenance of additional features of apical-basal polarity. |
|
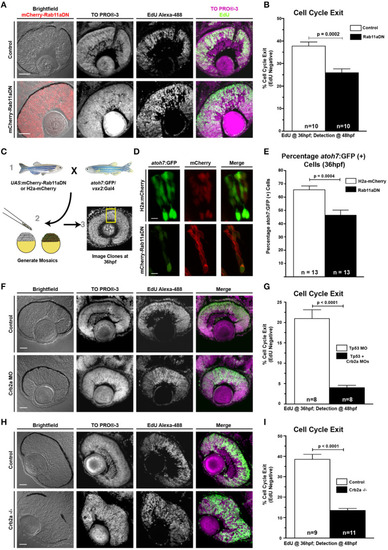
Rab11aDN expression and Crb2a loss-of-function promote RPC proliferation. |
|
Ectopic expression of the full-length Crb2a promotes RPC proliferation. |
|
Crb2aINT localized to Rab11a recycling endosomes and inhibition of recycling endosome activity promote RPC proliferation. |
|
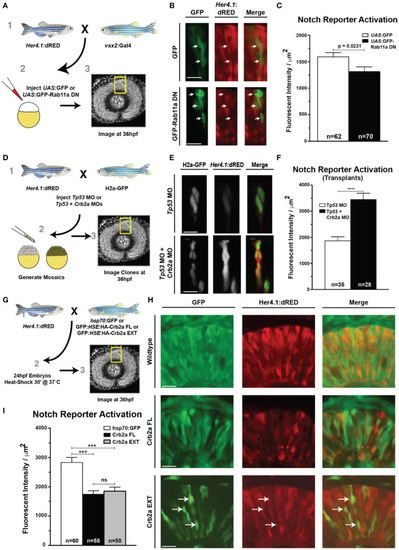
Rab11aDN expression and Crb2a function to inhibit Notch-reporter activation. |
|
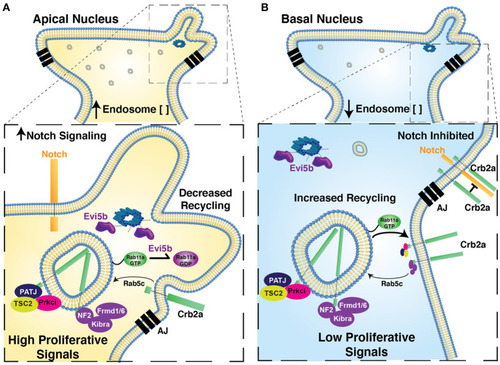
Proposed model for relationship of Nuclear Position, endosome activity, and the regulation of polarized signaling and retinal neurogenesis. Model figure suggesting the functional significance of nuclear position in the regulation of apical endosome concentration, Rab11a recycling endosome activity, the regulation of Crb2a apical protein localization and the regulation of autonomous signaling. In cells with apical nuclei |
|
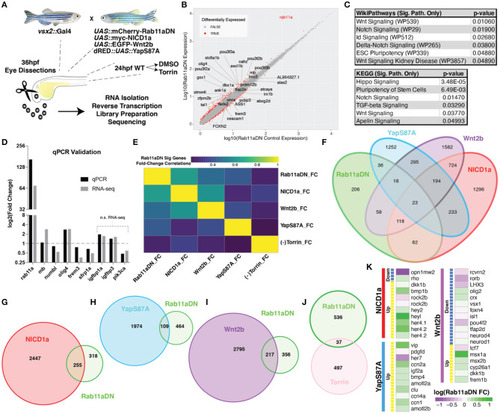
RNA-sequencing of Rab11aDN retinas identifies shared features with modulation of multiple signaling pathways control RPC proliferation. |