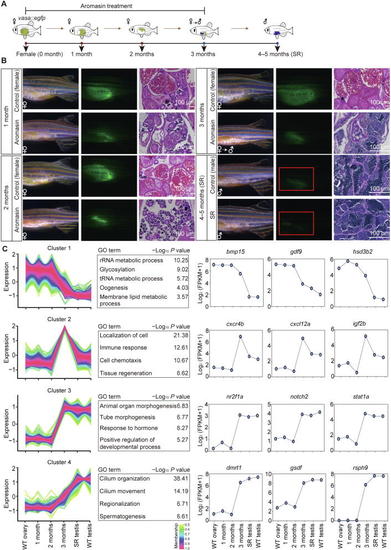
|
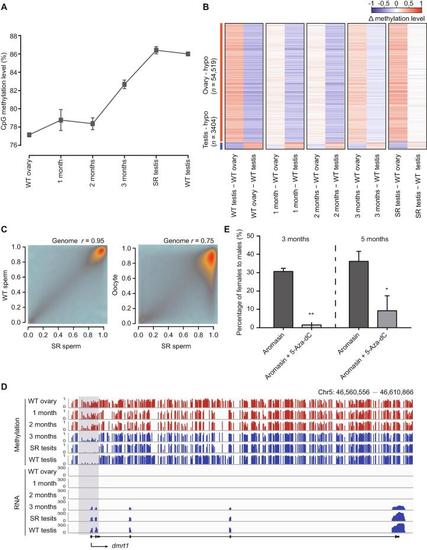
DNA methylomeisresetfrom the female pattern to the male pattern during zebrafish sex transition A. The global DNA methylation reprogramming during zebrafish sex transition. The methylation level was calculated as the average across all covered CpGs in the genome. The data are represented by mean ± SD. The samples include WT ovary, gonads from zebrafish treated with aromasin for 1 month, 2 months, or 3 months, SR testis, and WT testis. B. The dynamic changes for DMRs between zebrafish ovary and testis during zebrafish sex transition. The Δ methylation heatmap displays the difference in DNA methylation level between two consecutive stages during zebrafish sex transition. ‘n’ represents the number of ovary-specific hypo regions or testis-specific hypo regions. C. The correlation analysis of CpG methylation levels between SR sperm and WT sperm or oocyte. The Pearson correlation coefficients (r) is shown on the top. D. Snapshot of DNA methylation and gene expression tracks for the sex determination gene dmrt1 in WT ovary, gonads from zebrafish treated with aromasin for 1 month, 2 months, or 3 months, SR testis, and WT testis. The promoter of dmrt1 is defined as regions 1000 bp upstream and downstream of the TSS. Dynamic regions around the promoter are highlighted in gray. E. Sex ratio within populations at 3 months (aromasin only, n = 36; aromasin + 5-Aza-dC, n = 47) and 5 months (sex-reversed stage; aromasin only, n = 36; aromasin + 5-Aza-dC, n = 38) after drug treatment. Fish were treated with aromasin only / aromasin + 5-Aza-dC for 3 months, and then raised till 5 months. Three independent experiments were performed for each drug treatment group. The data are represented by mean ± SEM. Statistical significance was calculated by unpaired two-sided t-test. *, P < 0.05; **, P < 0.01. DMR, differentially methylated region; hypo, hypomethylated.
|