- Title
-
Spatiotemporal Characterization of Anterior Segment Mesenchyme Heterogeneity During Zebrafish Ocular Anterior Segment Development
- Authors
- Van Der Meulen, K.L., Vöcking, O., Weaver, M.L., Meshram, N.N., Famulski, J.K.
- Source
- Full text @ Front Cell Dev Biol
|
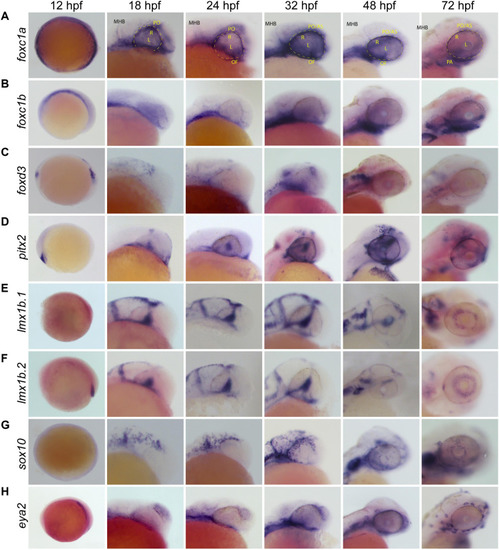
Whole Mount |
|
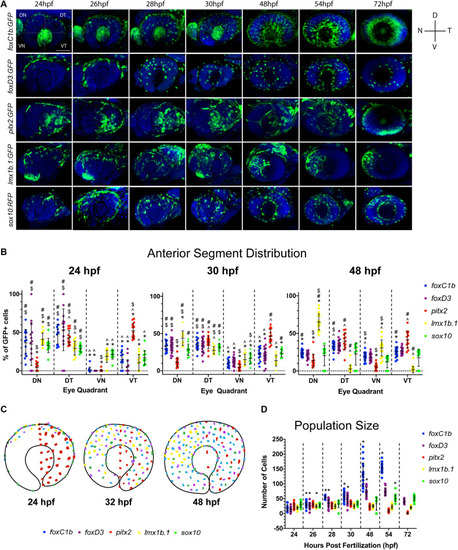
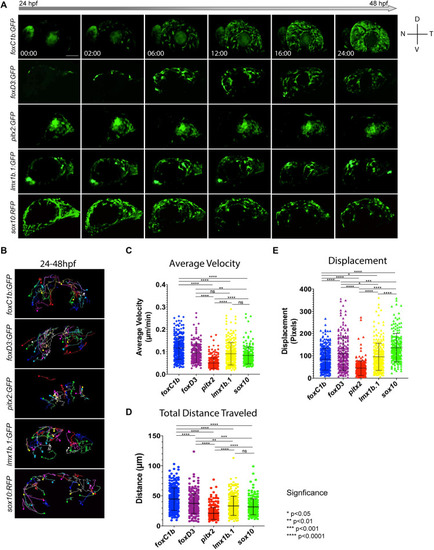
Two-color fluorescent |
|
Periocular mesenchyme subpopulation distribution analysis. |
|
|
|
Anterior segment mesenchyme single cell clustering analysis at 48 hpf. |
|
Gene expression of sequencing-derived genes. Whole-mount |