- Title
-
Single-molecule RNA detection at depth via hybridization chain reaction and tissue hydrogel embedding and clearing
- Authors
- Shah, S., Lubeck, E., Schwarzkopf, M., He, T.F., Greenbaum, A., Sohn, C.H., Lignell, A., Choi, H.M., Gradinaru, V., Pierce, N.A., Cai, L.
- Source
- Full text @ Development
|
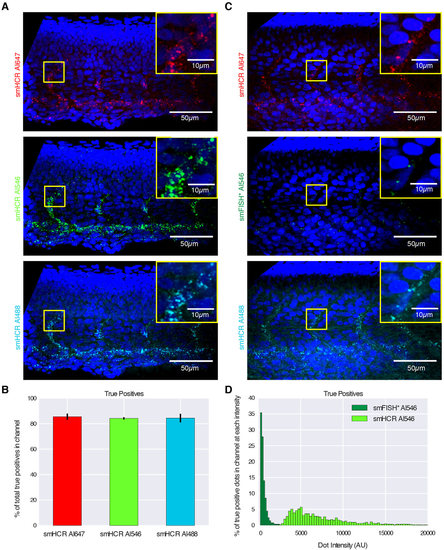
Imaging single mRNAs within whole-mount zebrafish embryos using smHCR. (A) Dot colocalization in three channels (DAPI in blue): smHCR (Alexa Fluor 647), smHCR (Alexa Fluor 546), smHCR (Alexa Fluor 488). (B) True positive rates for each channel in A (median±median absolute deviation, N=6 embryos). (C) Comparison of smHCR and smFISH* via dot colocalization in three channels: smHCR (Alexa Fluor 647), smFISH* (Alexa Fluor 546), smHCR (Alexa Fluor 488). Channel pairs between A and C are shown with the same contrast; Alexa Fluor 546 images illustrate the difference in intensity between amplified smHCR dots and unamplified smFISH* dots. (D) True positive dot intensities for smHCR (Alexa Fluor 546; N=6 embryos) and smFISH* (Alexa Fluor 546; N=3 embryos). Target mRNA: kdrl (expressed in the endothelial cells of blood vessels). Microscopy: spinning disk confocal. Probe sets: 39 probes per set, each addressing a 30 nt target subsequence. Embryos fixed: 27hpf. See Figs S6-S8 for additional data. |