- Title
-
A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P
- Authors
- Schenck, A., Bardoni, B., Moro, A., Bagni, C., and Mandel, J.L.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
FMRP interacts with CYFIP1/2. (A) Interaction in vitro between GST-tagged full-length FMRP and in vitro translated CYFIP1 N and C terminus (N, C), overlapping by 184 amino acids. GST-pd assays were performed in the presence of 100 mM sodium chloride. In each lane, 10% of the translation product used in each reaction or 40% of the eluate from glutathione beads was loaded. Both overlapping parts of CYFIP1 were retained by GST-FMRP but not by GST alone. Luciferase (L), the negative control, was bound neither by GST-FMRP nor by GST alone. (B) GST-pd assays using additional truncated constructs of CYFIP1 revealed that the N + C overlapping region is not exclusively responsible for interaction with FMRP. No binding affinity to GST-FMRP has been shown by construct 4 (amino acids 1-416). (C) Interaction of CYFIP1 with FMRP in vivo. FMRP, FXR1P, FXR2P, and nucleolin were coimmunoprecipitated from cytoplasmic lysate of HeLa cells by 8 μg of pAb no. 1467 raised against CYFIP1. A functional unrelated protein, lactate dehydrogenase, was not precipitated but was present in the cytoplasmic extract (lane 3). None of the proteins was precipitated by the same amounts of rabbit IgG, demonstrating the specificity of the experiment. One percent of each coimmunoprecipitation reaction was loaded to reveal CYFIP1 and the rabbit IgG heavy chain, eight percent to reveal coprecipitated proteins. (D) CYFIP2 is interacting with GST-FMRP in vitro. CYFIP1 translation product (amino acids 1-959, lane 1) and CYFIP2 translation product (amino acids 1-890, lane 2) were retained by GST-FMRP, but not the negative control (luciferase, lane 3). Assay and loading were performed as described in A. Proteins were separated in 8 or 10% SDS-polyacrylamide gels. |
|
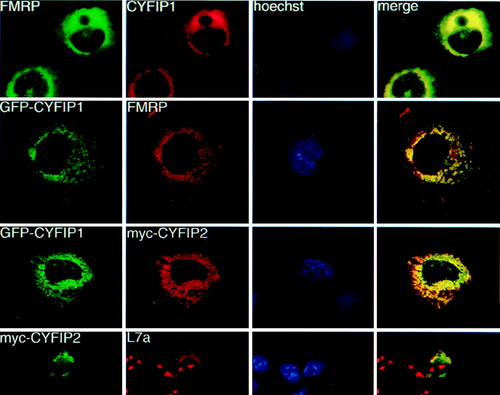
Coimmunolocalization studies in transiently transfected Cos cells. Row 1: cells transfected with CYFIP1 and FMRP iso7. FMRP was revealed by mAb 1C3, CYFIP1 by pAb no. 1465. Row 2: cells cotransfected with GFP-CYFIP1 and FMRP iso7, revealed by mAb 1C3. Row 3: cells cotransfected with GFP-tagged CYFIP1 and a myc-tagged CYFIP2 construct, revealed by mAb α-myc 9E10. Row 4: Cos transfected with myc-CYFIP2 (observed by 9E10) and compared with localization of endogenous ribosomes, revealed by a pAb raised against L7a, a protein of the ribosomal large subunit. Anti-L7a also stains nucleoli, the loci of ribosome assembly. Column 3: Hoechst staining. Column 4: merging images (yellow) from series 1 and 2, respectively, indicate colocalization. |
|
Subcellular fractionations. (A) CYFIP1/2, FMRP, and L7a content in three subcellular fractions of HeLa cells analyzed by Western blot by using pAb no. 1465, pAb no. 1716, mAb 1C3, and anti-L7a. Fractions represent nuclei (N), insoluble cytoplasm (P100), and soluble cytoplasm (S100). Signals from three independent experiments were quantified. Percentage of total cellular proteins in each fraction is indicated on the right. (B) CYFIP1/2 are present in synaptosomes. Western blot analysis of proteins in total brain extract (15 μg loaded) and synaptosomal extract (14 μg loaded). |
|
Selective interaction of CYFIP1/2 with FMRP, FXR1P, and FXR2P. (A) Interaction between the CYFIP proteins and the members of the FXR family, determined by yeast two-hybrid system. Diagram illustrating activation of the β-gal reporter in yeast coexpressing the following proteins (from Left to Right): FMRP + CYFIP1, FXR1P + CYFIP1, FXR2P + CYFIP1, FMRP + CYFIP2, FXR1P + CYFIP2, and FXR2P + CYFIP2, measured by liquid ONPG assay. At the foot of each bar, a X-gal filter lift assay is shown. Blue (black) color indicates β-gal activity induced by interaction of the two transformed fusion proteins. CYFIP2 interacts also with the FMRP-related proteins FXR1P/2P, whereas CYFIP1 interaction is restricted to FMRP. (B) The His6 pull-down assay (His-pd) assay using His6-tagged full-length CYFIP1 and in vitro translated proteins FMRP iso7 (lane 1), FXR1P iso a/P70 (lane 2), FXR2P (lane 3), and luciferase (lane 4). His-pd assays were performed in a buffer containing 200 mM sodium chloride. Loadings were performed as indicated in the legend to Fig. 1. FMRP bound specifically to CYFIP1, whereas FXR1P, FXR2P, and the negative control luciferase were not retained. None of the proteins is retained by beads incubated with extract from SF9 cells infected by wild-type baculovirus. |
|
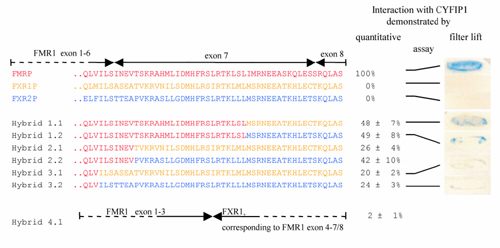
The FMRP sequence encoded by FMR1 exon 7 is implicated in the interaction with CYFIP1. Yeast two-hybrid assay between CYFIP1 and chimeric FMRP/FXR1P and FMRP/FXR2P proteins. In hybrid constructs, 1.1-2.2 increasing parts of FMRP exon 7 were replaced by FXR1P/2P, in hybrids 3.1 and 3.2 the entire exon 7. In construct 4.1, exon 4-7/8 of FMR1 is substituted by the FXR1 sequence. To the right of each construct, the strength of interaction with CYFIP1 in relation to interaction FMRP-CYFIP1 is indicated. For constructs 1.1, 1.2, 3.1, and 3.2, also an X-gal filter lift assay is shown. A more detailed representation of the hybrid constructs, the FMRP exon 7 region, and the corresponding FXR1P/2P sequences is published as supplemental data on the PNAS web site, www.pnas.org. |
|
The FMRP sequence encoded by FMR1 exon 7 is implicated in the interaction with CYFIP1. Yeast two-hybrid assay between CYFIP1 and chimeric FMRP/FXR1P and FMRP/FXR2P proteins. FMRP sequences are in red, FXR1P in yellow, and FXR2P in blue. In hybrid constructs 1.1–2.2, increasing parts of FMRP exon 7 were replaced by FXR1P or -2P, in hybrids 3.1 and 3.2, the entire exon 7. In construct 4.1, exon 4-7/8 of FMR1 is substituted by the FXR1 sequence. On the right of each construct, the strength of interaction with CYFIP1 in relation to interaction FMRP--CYFIP1 is indicated. For constructs 1.1, 1.2, 3.1, and 3.2, a 5-bromo-4-chloro-3-indolyl β-D-galactoside filter lift assay is shown. |