- Title
-
Highly efficient germ-line transmission of proviral insertions in zebrafish
- Authors
- Gaiano, N., Allende, M., Amsterdam, A., Kawakami, K., and Hopkins, N.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
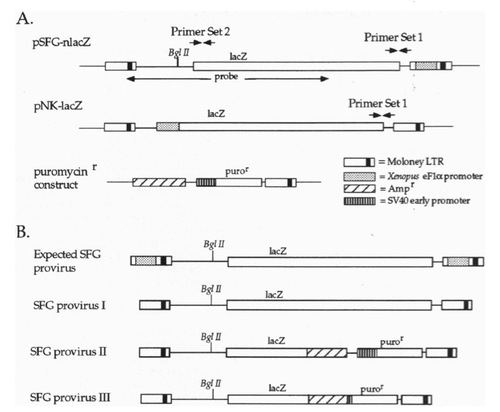
Schematic representation of the plasmids used and of the predicted and observed proviral structures. (A) Maps of SFG (with 3′-LTR modified to contain the Xenopus eFlα promoter), NK, and the construct used to confer resistance to puromycin. The locations of primer sets 1 and 2 are indicated, as is the region of SFG that was used as a probe for Southern blots. (B) The expected SFG proviral genome, and the actual SFG proviral genomic structures. |
|
Southern blot analysis of DNA from transgenic F1 progeny of founder SFG77. The genomic DNA was digested with BglII and probed with the sequence indicated in Fig. 1A. Each insertion is expected to produce two junction fragments with sizes characteristic of the site of integration. The result of segregation during meiosis is apparent in lanes 2-7 and 10-12. For example, the Fl represented in lane 11 has four bands representing two insertions. These two insertions can be seen independently in the Fis represented in lanes 10 and 12. |