- Title
-
Combined genetic and chemical screens indicate protective potential for EGFR inhibition to cardiomyocytes under hypoxia
- Authors
- Heliste, J., Jokilammi, A., Vaparanta, K., Paatero, I., Elenius, K.
- Source
- Full text @ Sci. Rep.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
shRNA library screening to identify modulators of cardiomyocyte survival. ( |
|
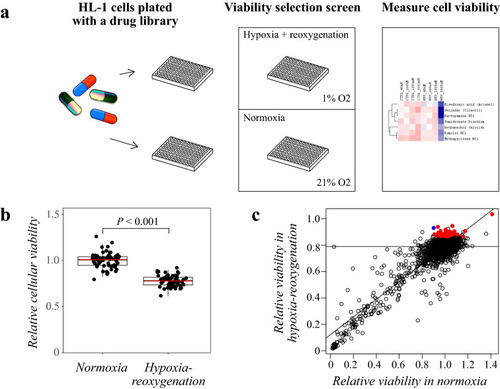
Drug library screening to identify compounds affecting cardiomyocyte viability. Schematic overview of the drug library screen. HL-1 cells were plated with a drug library consisting of 689 FDA-approved compounds. Cells were exposed to hypoxia-reoxygenation for a viability dropout screen. As the readout, cellular viability was measured with CTG assays. ( |
|
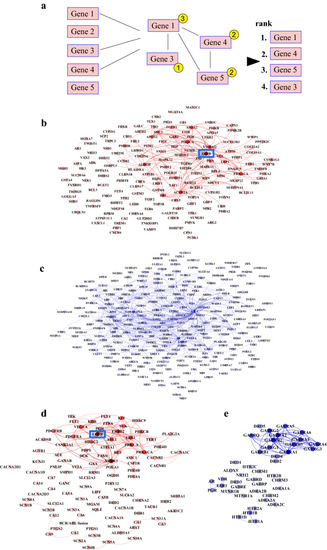
Protein–protein interaction analyses of significant hits from shRNA and drug library screens. ( |
|
Effects of gefitinib on cardiomyocytes and zebrafish embryos challenged with hypoxia-reoxygenation. ( PHENOTYPE:
|
|
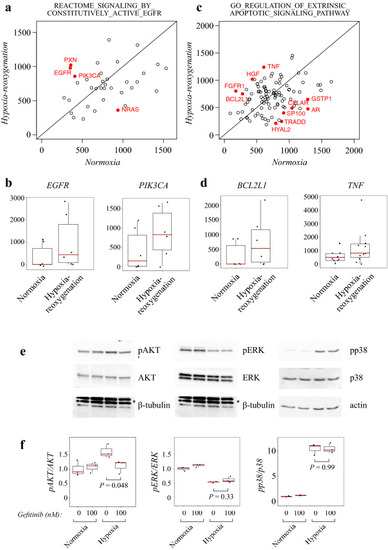
Pathways involved in hypoxia-reoxygenation response and EGFR-mediated signal transduction. ( |