- Title
-
Integrin intra-heterodimer affinity inversely correlates with integrin activatability
- Authors
- Sun, G., Guillon, E., Holley, S.A.
- Source
- Full text @ Cell Rep.
|
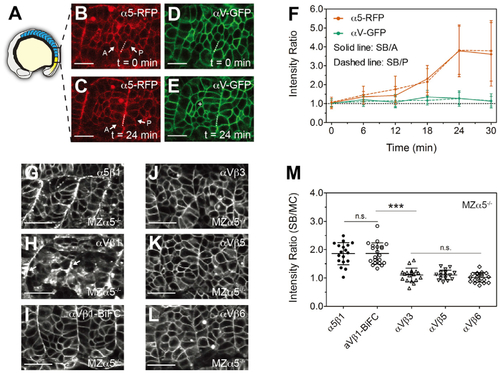
(A) Illustration of a zebrafish embryo highlighting the somites (blue) and presomitic mesoderm (yellow). (B?E) Confocal images of integrin ?5-RFP (B and C) and ?V-GFP (D and E) in wild-type (WT) embryos. As the somite boundary (SB) forms, ?5 clusters to the basal side (dashed lines) of the anterior (A) and posterior (P) boundary cells (arrows). The white cross in (E) denotes a mesenchymal cell (MC) within a somite. Scale bars, 20 ?m. (F) Basal/apical ratio of integrin intensity in anterior (SB/A, solid line) and posterior (SB/P, dashed line) boundary cells. Data are mean ± SD from n = 15 cell pairs in six embryos. (G?L) Integrin ?5-Aquamarine (Aqm) and ?V-Aqm co-expressed with different integrin ? subunits tagged with mCitrine (mCit) in developing somites of MZ?5?/? embryos. (G) ?5?1, (H) ?V?1, (I) ?V?1-BiFC (bimolecular fluorescence complementation, used to increase heterodimer stability), (J) ?V?3, (K) ?V?5, and (L) ?V?6. Arrows in (H) indicate clustering on the somite border. Scale bars, 30??m. (M) Clustering quantification via the SB/MC intensity ratio. Details of ROI selection shown in Figure S1A. ?5?1, n = 18 measurements (12 embryos); ?V?1-BiFC, n = 21 (13 embryos); ?V?3, n = 18 (8 embryos); ?V?5, n = 16 (14 embryos); ?V?6, n = 19 (9 embryos). Data are mean ± SD. ???p < 0.0001; n.s., not significant (two-sided t test). See also Figure S1. |
|
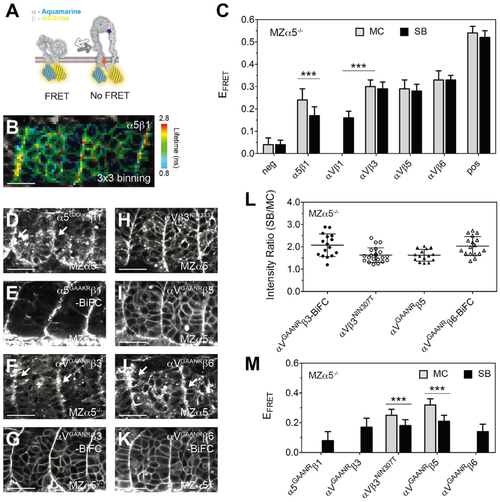
(A) Illustration of the FRET assay for the integrin conformation. The integrin ? subunit cytoplasmic tail was tagged with Aqm as a FRET donor, and the ? subunit was tagged with mCit as a FRET acceptor. When the cytoplasmic tails separate during integrin activation, FRET should be reduced. The locations of the ?GAAXR mutations (orange star) and the ?3NIN333T mutation (purple star) are indicated. (B) Heatmap of the fluorescence lifetime of integrin ?5-Aqm co-expressed with ?1-mCit (denoted ?5?1). The raw image and lifetime distribution are shown in Figures S1C and S1D. Warm colors on the SB represent longer donor lifetimes, indicating weaker FRET and the active conformation. (C) FRET efficiency (EFRET) of different integrin heterodimers. Sample size is the same as in Figure 1M, except ?V?1, n = 20 measurements (12 embryos). (D?K) Activatable integrin alleles: ?5GAAKR-Aqm co-expressed with ?1-mCit (D), ?5GAAKR?1-BiFC (E), ?VGAANR-Aqm co-expressed with ?3-mCit (F), ?VGAANR?3-BiFC (G), ?V-Aqm co-expressed with N-glycan wedge allele ?3NIN333T-mCit (H), ?VGAANR-Aqm co-expressed with ?5-mCit (I), ?VGAANR-Aqm co-expressed with ?6-mCit (J), and ?VGAANR?6-BiFC (K). White arrows in (D), (F), and (J) indicate clustering on the somite border. Scale bars, 30??m. (L) Clustering quantification of activatable integrin alleles by the SB/MC intensity ratio. ?VGAANR?3-BiFC, n = 16 (9 embryos); ?V?3NIN333T, n = 21 (15 embryos); ?VGAANR?5, n = 15 (8 embryos); ?VGAANR?6-BiFC, n = 17 (8 embryos). ?5GAAKR?1-BiFC clustering cannot be measured because of the poor membrane expression in the mesenchyme. (M) EFRET of activatable integrin alleles. Sample size is the same as in (L), except ?5GAAKR?1, n = 18 (9 embryos); ?VGAANR?3, n = 14 (9 embryos); and ?VGAANR?6, n = 15 (7 embryos). EFRET of ?V?1 (C), ?5GAAKR?1, ?VGAANR?3, and ?VGAANR?6 (M) cannot be measured in the MC because of the poor membrane expression. (C and M) Data are mean ± SD. ???p < 0.0001, two-sided t test. All experiments are in MZ?5?/? embryos. |
|
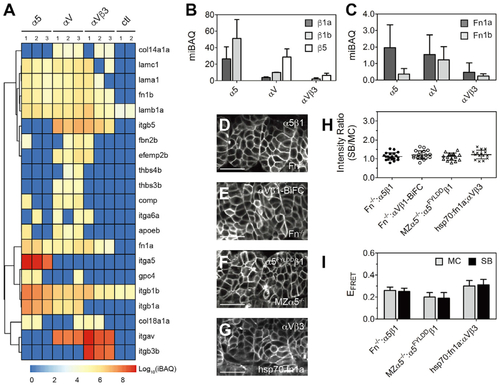
(A) Integrins and ECM proteins co-immunoprecipitated with integrins ?5, ?V, or ?V?3 identified via mass spectroscopy. The intensity-based absolute quantification (iBAQ) from each replicate is color coded to show relative protein abundance. Hierarchical cluster analysis is shown as the dendrogram (see Tables S2 and S3 for protein names). Note that basement membrane ligand laminins (lama1, lamb1a, lamc1) are roughly equal in all three datasets, while thrombospondins (thbs3b, thbs4b) and cartilage oligomeric matrix protein (comp/thbs5) are found exclusively in the ?V dataset. ctl, control (FLAG-tagged myristoylated membrane-anchored GFP [mem-GFP]). (B and C) Integrin ? subunits (B) and Fn (C) quantification using median-normalized iBAQ (miBAQ). Bar indicates mean ± SD, n = 3. (D?G) Somite localization of integrin ?5?1 (D) and ?V?1-BiFC (E) in Fn double-mutant Fn?/? (fn1a?/?;fn1b?/?) embryos, ligand binding-deficient ?5FYLDD?1 in MZ?5?/? embryos (F), and ?V?3 in heat shock promoter-driven Fn1a-mKikumi transgenic (hsp70:fn1a) embryos (G). Scale bars, 30 ?m. (H and I) Clustering quantification (H) and EFRET (I) of ?5?1 in the absence of Fn, ?5FYLDD ?1 in MZ?5?/? embryos, and ?V?3 exposed to extra Fn1a. Fn?/?: ?5?1, n = 20 measurements (9 embryos); Fn?/?: ?V?1-BiFC, n = 19 (11 embryos); MZ?5?/?: ?5FYLDD ?1, n = 16 (8 embryos); hsp70:fn1a; ?V?3, n = 15 (7 embryos). Data are mean ± SD. |
|
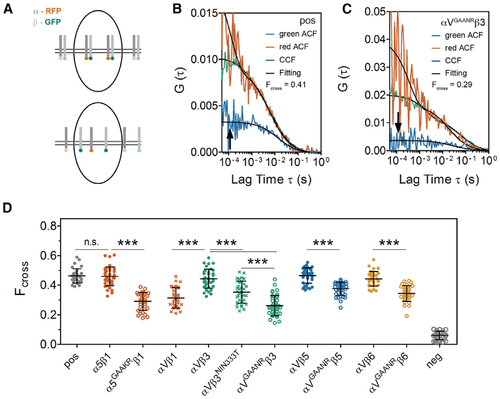
(A) Illustration of fluorescence cross-correlation spectroscopy (FCCS) measurements. The integrin ? subunit cytoplasmic tail was tagged with RFP and the ? subunit was tagged with GFP. When the two subunits move together through the confocal volume (upper panel), the green and red intensity fluctuations correlate, leading to a high cross-correlation curve (arrow in B); conversely, when the heterodimer subunits dissociate (lower panel), there is a lower cross-correlation curve (arrow in C). (B and C) FCCS measurements of the positive control (pos), which is a mem-GFP-RFP tandem fusion (B) and FCCS measure of ?VGAANR?3 (C). The auto-correlation functions (ACFs) for each channel are shown in red and green while the cross-correlation between the two channels is in blue. Data fitting is shown in black. Measurements were performed on the cell surface in somite MCs (white cross in Figure 1E). (D) Fcross of different integrin heterodimers calculated from FCCS. A lower Fcross indicates a weaker intra-heterodimer association. pos, positive control, mem-GFP-RFP tandem; neg, negative control, co-expression of mem-GFP and mem-RFP. Data are mean ± SD. ???p < 0.0001, n.s., not significant (two-sided t test). |