- Title
-
Fishing for cures: The alLURE of using zebrafish to develop precision oncology therapies
- Authors
- Astone, M., Dankert, E.N., Alam, S.K., Hoeppner, L.H.
- Source
- Full text @ NPJ Precis Oncol
|
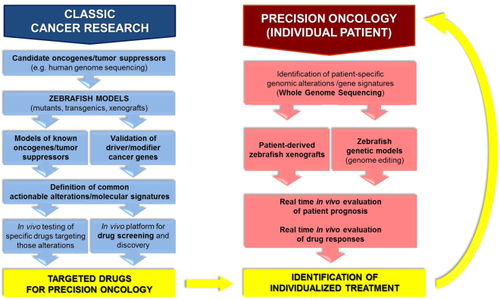
Applications of the zebrafish model in precision oncology. Classic cancer research using zebrafish has contributed to precision oncology through the establishment of numerous cancer models, leading not only to significant advancements in cancer biology, but also to the definition of targeted drugs suitable for personalized cancer treatments (blue, left). Possible applications of zebrafish in the clinic to drive personalized therapies for specific patients have also been shown. The feasibility of this approach has been demonstrated through the use of patient-derived zebrafish xenografts and generation of transgenic zebrafish modeling mutations or translocations defining a specific patient’s tumor (red, right) |
|
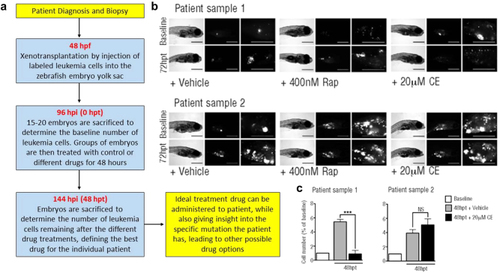
Precision oncology approach to leukemia drug screening using zebrafish. |
|
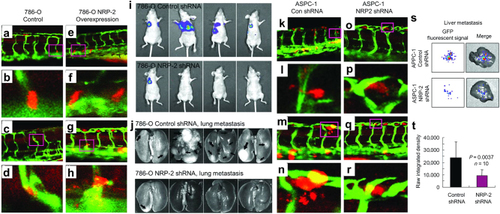
Human cancer cell xenograft models of extravasation in zebrafish and metastasis in mice. |