- Title
-
Wnt/?-Catenin Signaling Defines Organizing Centers that Orchestrate Growth and Differentiation of the Regenerating Zebrafish Caudal Fin
- Authors
- Wehner, D., Cizelsky, W., Vasudevaro, M.D., Özhan, G., Haase, C., Kagermeier-Schenk, B., Röder, A., Dorsky, R.I., Moro, E., Argenton, F., Kühl, M., and Weidinger, G.
- Source
- Full text @ Cell Rep.
|
Wnt/β-Catenin Pathway Activation during Zebrafish Caudal Fin Regeneration (A) Cartoons summarizing relevant anatomical structures and expression domains of a fin regenerate during the outgrowth phase. Whole-mount and longitudinal section views are shown. (B) mCherry RNA expression in 7xTCF:mCherry transgenic regenerates. Note transcript expression at 12 hpa in the interray tissue (arrowheads) and at 24 hpa distally to the bony ray (arrow). (C) mCherry expression is confined to the mesenchyme at all stages analyzed. The basal epidermal layer is devoid of staining (asterisk). (D) axin2 (green) and mCherry RNA (red) are coexpressed in the distal blastema (arrowhead) and interray cells (arrow) in 7xTCF:mCherry transgenic regenerates. (E) gfp RNA expression in 6xTCF:dGFP transgenic regenerates is confined to the blastema. The basal epidermal layer is devoid of staining (asterisk). (F) lrp5 transcripts are confined to proximal mediolateral domains of the blastema and are absent in the distal (arrowhead) blastema, the osteoblast progenitors (asterisk), and the basal epidermal layer (arrow). lrp6 transcripts are predominantly detected in the distal blastema (arrowhead). Asterisk: (pre)osteoblasts; arrow: basal epidermal layer. (G) RT-PCR of indicated genes on cDNA derived from fluorescence-activated-cell-sorted 7xTCF:mCherry; her4.3:EGFP double transgenic regenerates at 72 hpa. Endogenous lrp5/6 transcripts are not detected in the fluorescence-negative, blastema-free cell fraction that contains epidermal cells whereas spry4 is. In (B)?(G), confocal images of a whole-mount (D) or longitudinal section (E) are shown. Black arrowheads, amputation plane. Scale bars, 200 μm, 100 μm (D, whole mount),10 μm (D), and 100 μm (sections). |
|
β-Catenin Pathway Activity Is Detected in Different Blastemal Compartments with Distinct Proliferative Properties (A) gfp transcripts (green) in 6xTCF:dGFP transgenic regenerates are strongly detected in cells of the distal blastema and in cells medially to the Runx2+ osteoblast progenitors (red; asterisk). gfp transcripts are only weakly detected in Runx2+ preosteoblasts (arrowheads). (B) aldh1a2 (green) and mCherry RNA (red) are coexpressed in the distal-most blastema in 7xTCF:mCherry transgenic regenerates. (C) gfp RNA in 6xTCF:dGFP regenerates is detected in the distal-most, Pcna-negative blastema (arrow) and in a subpopulation of the Pcna+, proximal blastema (arrowheads). (D) gfp is coexpressed with and1 in the actinotrichia forming cells of 6xTCF:dGFP transgenics. Asterisk: (pre)osteoblasts. Note that the absence of Wnt reporter activity in the (pre)osteoblasts is due to a less sensitive staining procedure. In (A)?(D), confocal images of longitudinal sections (A and C?D) or whole mounts (B). Scale bars, 100 μm and 10 μm (whole mount) and 100 μm (sections). |
|
Wnt/β-Catenin Signaling Must Be Inhibited in the Distal and Proximal Blastema for Regenerative Growth to Be Stalled (A) Cartoon showing strategy for tissue-specific inducible overexpression of the Wnt/β-catenin signaling inhibitor axin1 using the TetON system. (B) ubiquitin promoter-driven axin1 overexpression in the epidermis and the blastema is sufficient to block regenerative growth. (C) keratin4 promoter-driven axin1 overexpression in the epidermis excluding the basal layer does not interfere with regenerative growth. (D) keratin18 promoter-driven axin1 overexpression in the basal epidermal layer (and ectopic expression in the proximal medial blastema) does not interfere with regenerative growth. (E) axin1 overexpression in the committed sp7+ osteoblasts does not interfere with regenerative growth. (F) axin1 overexpression in the medial proximal blastema has only little impact on regenerative growth in her4.3:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish. White arrowhead: distal-most blastema. In (B)?(F), BF, bright field. Small arrowheads, amputation plane. Scale bars, 200 μm (whole mounts); 100 μm (sections). Error bars indicate error of the mean. |
|
Wnt/β-Catenin Signaling in the Proximal Medial Blastema Is Required for Bone Formation (A) axin1 overexpression in the medial proximal blastema does not significantly reduce H3P+ blastemal cells in her4.3:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish. In contrast, proliferation is strongly reduced when axin1 is directed to both the distal and medial proximal blastema using the ubiquitin promoter. (B) axin1 overexpression in the medial proximal blastema but not in committed osteoblasts reduces the fraction of the regenerate stained with the bone calcification marker Alizarin Red. Bracket indicates the noncalcified distal region of the regenerate. (C) axin1 overexpression in the proximal blastema reduces expression of the osteoblast commitment marker sp7. In (A)-(C), arrowheads, amputation plane. Scale bar, 200 μm. Error bars indicate error of the mean. |
|
Wnt/β-Catenin Signaling Regulates Many Other Signaling Pathways (A) Eigengene values of three correlated WGCNA modules, representing the average relative expression values of all genes in the module, in ten experimental conditions. Values shown are the average of triplicate samples relative to 0 hpa wild-type samples. Error bars indicate error of the mean. (B) Cartoon of genes found in module 26 with genes annotated to be associated with Wnt, Fgf, and Hh signaling highlighted. (C) Samples in (D)?(M) were either heat shocked at 42 hpa (blue) or 66 hpa (green) and harvested 6 hr later. (D?M) Inhibition of Wnt/β-catenin signaling via axin1 or dkk1 overexpression interferes with pathway activity or expression of ligands, pathway components, and target genes of Hh (D), Bmp (E), RA (F), Igf (G), Notch (I and J), and Fgf (K) signaling. Expression of msxB and actβAa but not actβB remains unaffected using the same heat shock protocol (L and M). (H) axin1 overexpression for 6 hr reduces Igf1r phosphorylation as detected in western blots of whole regenerates at 72 hpa. (I) axin1 overexpression interferes with Notch signaling-dependent her4.3 promoter activity in her4.3:TetA AmCyan; hs:Axin1 double transgenic fish. In (D)?(M), scale bars, 200 μm. |
|
Wnt/β-Catenin Signaling Regulates Blastema Proliferation via Hedgehog and Retinoic Acid Signaling (A) noggin3 overexpression reduces lef1 (blue) but not mCherry (brown) expression in 7xTCF:mCherry; hs:nog3 double transgenic regenerates. (B) Overexpression of a dominant negative fgfr1 reduces lef1 (blue) but not mCherry (brown) expression in 7xTCF:mCherry; hs:dnfgfr1 double transgenic regenerates. (C) axin1 overexpression inhibits expression of the Fgf targets pea3 and erm, which is rescued by concomitant activation of Fgf/Ras signaling via overexpression of v-ras. (D) axin1 overexpression inhibits expression of the Hh target ptc2, which is rescued by concomitant activation of Hh signaling using the pharmacological compound SAG. (E) axin1 overexpression reduces the number of H3P+ cells in the blastema, which is rescued by concomitant activation of Hh signaling using SAG. (F) Inhibition of Hh signaling through exposure to cyclopamine reduces the number of H3P+ cells in the blastema, which is not rescued by concomitant activation of Wnt signaling following wnt8 overexpression. (G) Administration of RA via intraperitoneal injection rescues the reduction of H3P+ blastemal cell numbers caused by axin1 overexpression. In (A)?(G), arrowheads indicate amputation plane. Scale bars, 200 μm. Error bars indicate SEM. |
|
Wnt/β-catenin pathway activation, Tcf/Lef transcription factor and Lrp5/6 Wnt coreceptor expression during caudal fin regeneration, related to figure 1. (A) mCherry RNA expression in whole mounts of 7xTCF:mCherry transgenic regenerates. Note that transcript expression is detected at 6 hpa in the interray tissue (white arrowhead). (B) 7xTCF:mCherry reporter activity in the interray tissue (white arrowhead) is abolished upon dkk1 overexpression in 7xTCF:mCherry; hs:dkk1 double transgenic regenerates 6 hours post heat shock. (C) Confocal image showing mCherry fluorescence and Lef1 protein on sections of 7xTCF:mCherry transgenic regenerates. Note that mCherry fluorescence is not detected in the Lef1-positive basal epidermal layer. (D) mCherry fluorescence in 7xTCF:mCherry transgenic regenerates at different stages after amputation. Note that mCherry fluorescence is first detected in the interray tissue at 16 hpa. (E) Downregulation of axin2 expression in hs:Axin1 transgenic fish 6 hours post heat shock. (F) gfp RNA expression in Top:dGFP transgenic regenerates. Note that transcript expression is detected in the blastemal mesenchyme while the basal epidermal layer is devoid of signal (asterisk). (G) lef1, tcf1 (tcf7), tcf3a (tcf7l1a), tcf3b (tcf7l1b) and tcf4 (tcf7l2) are expressed in the regenerating fin. Hybridization with sense control probes revealed no signal. (H) Sections of whole mount stained regenerates shown in (G) reveal lef1 expression in the distal blastema (arrowhead) and the basal epidermal layer, tcf1 expression in the basal epidermal layer, lateral (asterisk) and distal blastema (arrowhead), tcf3a, tcf3b and tcf4 expression in the medial and lateral (asterisk) blastema. (I) tcf1 (brown) and lef1 (blue) are co-expressed in the epidermis of the regenerating fin. 5 (J) Dual color ISH of Tcf/Lef transcription factors and mCherry in 7xTCF:mCherry transgenic regenerates. tcf1, lef1 and tcf3a (blue) are co-expressed with mCherry (brown) in the distal blastema (black arrowheads), while tcf3b and tcf4 (blue) are not expressed in the distal mCherry (brown)-positive domain (white arrowheads). (K) lef1 expression is reduced in hs:Axin1 transgenic regenerates 6 hours post heat shock. (L) lrp5 and lrp6 are expressed in the regenerating fin. Hybridization with sense control probes revealed no signal. (M) Confocal image showing mCherry-positive and EGFP-positive cells on sections of 7xTCF:mCherry; her4.3:EGFP double transgenic regenerates at 72 hpa. (N) wnt8 overexpression does not cause ectopic Wnt reporter activation in the wound epidermis in 7xTCF:mCherry; hs:wnt8 double transgenic fish. (O) dkk1 overexpression interferes with mCherry (arrow) but not epidermal lef1 (asterisk) expression 3 hours after a single heat shock in 7xTCF:mCherry; hs:dkk1 double transgenic regenerates. (A-O) Small arrowheads: amputation plane. Scale bars: whole mounts, 200 μm and 100 μm (J); sections, 100 μm. |
|
Wnt/β-catenin signaling is active in cells of the distal-most scarcely proliferative blastema, which retain their relative localization during regenerative growth, plus in different domains of the proximal blastema, related to figure 2. (A) Reporter activity in proximally located blastemal regions is robustly observed in 6xTCF:dGFP transgenic regenerates (big white arrowhead), but is detected in 7xTCF:mCherry transgenic regenerates only in a minority of samples after prolonged staining (big white arrowhead, 1/8 regenerates). (B) axin2 expression is strongly detected in the distal blastema (arrow) plus weakly in the lateral blastema containing the actinotrichia-forming cells (arrowhead) and the (pre)osteoblasts (asterisk). 7 (C) aldh1a2 (blue) co-localize with mCherry (brown) transcripts in the distal-most blastema in 7xTCF:mCherry transgenic regenerates. Note that samples were stained for mCherry expression only shortly, so that only the strongest expression domain in the distal blastema was detected. (D) mCherry transcripts (brown) are detected distally to egfp (blue) transcripts in 7xTCF:mCherry; her4.3:EGFP double transgenic regenerates. (E) mCherry fluorescence is detected distally to the EGFP fluorescence-positive proximal blastema (asterisk) in 7xTCF:mCherry; her4.3:EGFP double transgenic regenerates. Optical section through the center of a whole mount blastema is shown. (F) 7xTCF:mCherry reporter activity is detected in the Pcna-negative distal-most blastema. Optical section through the center of a whole mount blastema stained for mCherry transcripts (short staining) and Pcna protein is shown. (G-H) Kaede fluorescence (G) and transcripts (H) in 7xTCF:3xKaede transgenic regenerates. (I-J) Kaede-positive Wnt-receiving cells located in the distal-most row of mesenchymal cells of a 3 day-old 7xTCF:3xKaede transgenic blastema were photoconverted from green to red fluorescence and traced for two consecutive days. Cells containing the converted Kaede (red) were still detected in the distal-most cell row 24 hours and 48 hours post conversion (arrowheads in I) despite the substantial regenerative growth that occurred in these fins (J). (A-J) Small arrowheads: amputation plane. Scale bars: whole mounts, 200 μm (A, G-H, J) and 100 μm (C-F, I); sections, 100 μm. |
|
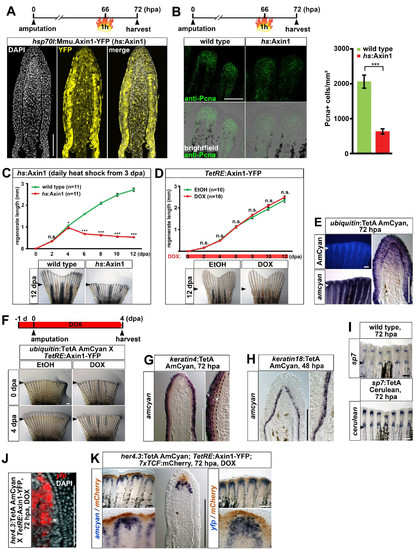
Characterization of a panel of transgenic TetON driver lines that allow for tissuespecific inducible gene expression in the adult caudal fin, related to figure 3. (A) Confocal images of YFP fluorescence in longitudinal sections of hsp70l:Mmu.Axin1-YFP (in short hs:Axin1) transgenic regenerates 6 hours post heat shock. Note that expression is ubiquitous, although levels differ greatly between cells. (B) Systemic overexpression of axin1 reduces the number of Pcna-positive cells in hs:Axin1 transgenic regenerates. A single optical section through the mesenchyme of a whole mount regenerate is shown. Error bars indicate error of the mean. 9 (C) Overexpression of axin1 starting at 3 dpa strongly interferes with regenerative growth in hs:Axin1 transgenic fish. (D) DOX treatment starting immediately after amputation does not affect regenerative growth in TetRE:Axin1-YFP transgenic fish compared to EtOH-treated controls. (E) AmCyan fluorescence and transcripts in ubiquitin:TetA AmCyan transgenic regenerates. Note that expression is fairly ubiquitous in the epidermis and the distal blastema, and covers a substantial part of the proximal blastema. (F) DOX treatment for 5 days starting 1 day before amputation interferes with regeneration in ubiquitin:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish. (G) amcyan RNA expression in keratin4:TetA AmCyan transgenic regenerates is detected in the epidermis excluding the basal epidermal layer (asterisk). (H) amcyan RNA expression in keratin18:TetA AmCyan transgenic regenerates is confined to the basal epidermal layer. (I) cerulean RNA expression recapitulates endogenous sp7 expression in sp7:TetA Cerulean transgenic regenerates. (J) yfp transcripts are detected medially to the osteoblast progenitors (asterisk) in her4.3:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish treated with DOX for 8 hours. Confocal image of a longitudinal sections is shown. (K) amcyan and yfp transcripts (blue) are detected proximally to mCherry (brown) in regenerates of her4.3:TetA AmCyan; TetRE:Axin1-YFP; 7xTCF:mCherry triple transgenic fish treated with DOX for 12 hours. (A-K) Small arrowheads: amputation plane. Scale bars: whole mounts, 500 μm (F) and 200 μm (B, E, I, K); sections, 100 μm. |
|
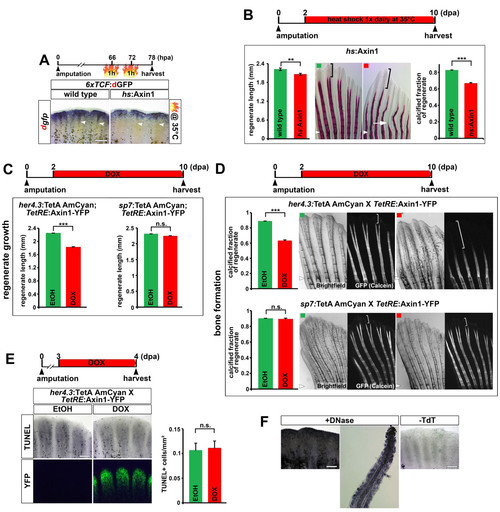
Inhibition of Wnt/β-catenin signaling in the proximal medial blastema interferes with regenerative growth and bone calcification, related to figure 4. (A) Mild heat shocks at 35°C strongly reduce proximal 6xTCF:dGFP Wnt reporter activity (arrowhead) but have little impact on distal Wnt reporter expression in 6xTCF:dGFP; hs:Axin1 double transgenic fish. (B) Systemic overexpression of axin1 at a level that has little impact on regenerative growth (daily heat shock at 35°C), strongly reduces the fraction of the regenerate containing calcified bones in hs:Axin1 transgenic fish. Bracket indicates the non-calcified distal region of the regenerate. Note that calcified bones are also thinner and shorter (arrow) compared to wild type control. (C) axin1 overexpression in the proximal medial blastema for 8 days reduces regenerate length in her4.3:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish treated with DOX. axin1 overexpression in committed osteoblasts for 8 days has no effect on regenerate length in sp7:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish treated with DOX. n = 9 fish, 6 rays each. (D) axin1 overexpression in the proximal medial blastema for 8 days in her4.3:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish treated with DOX strongly reduces the fraction of the 11 regenerate containing calcified bones as determined by Calcein staining. axin1 overexpression in committed osteoblasts has no effect on bone calcification in sp7:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish treated with DOX. Bracket indicates the non-calcified distal region of the regenerate. n = 9 fish, 6 rays each. (E) axin1 overexpression in the proximal medial blastema for 1 day does not enhance cellular apoptosis as detected by TUNEL assay in her4.3:TetA AmCyan; TetRE:Axin1-YFP double transgenic fish treated with DOX. (F) Positive and negative control for the TUNEL assay shown in (E). Longitudinal section of the DNAseI-treated positive control shows that TUNEL+ cells can be detected in the entire fin regenerate, including the osteoblasts. (B, D) Small arrowheads: amputation plane. (A, E-F) Scale bars: 200 μm. (B-E) Error bars indicate error of the mean. |
|
Wnt/β-catenin signaling receives little reciprocal input from other developmental signaling pathways during fin regeneration, related to figure 5. (A) sost and wnt10a expression is strongly reduced upon axin1 overexpression for 6 hours in hs:Axin1 transgenic regenerates. (B) bmp4 (blue) and mCherry (brown) transcripts co-localize in the distal-most blastema (white arrowhead) in 7xTCF:mCherry transgenic regenerates. Note that samples were stained for mCherry 13 expression only shortly, so that only the strongest expression domain in the distal blastema was detected. (C) actβB (blue) and mCherry (brown) transcripts co-localize in the distal most blastema in 7xTCF:mCherry transgenic regenerates (arrowhead). Note that samples were stained for mCherry expression only shortly, so that only the strongest expression domain in the distal blastema was detected. (D-H) 7xTCF:mCherry reporter expression is robustly detectable in severely reduced regenerates obtained after prolonged inhibition of Igf (D), Fgf (E), Notch (F), Activin (G) or RA (H) signaling for 48 hours or 72 hours using pharmacological compounds or heat shock-induced overexpression of pathway inhibitors. Pharmacological compounds: cyclopamine, Smoothened antagonist ? inhibitor of Hh signaling; IWR-1, Axin stabilizer ? Inhibitor of Wnt/β-catenin signaling; LY411575, γSecretase inhibitor ? inhibitor of Notch signaling; NVP-AEW541, Igf1 receptor inhibitor ? inhibitor of Igf signaling; SB431542, Activin receptor-like kinase receptor 4 (Alk4) inhibitor ? inhibitor of Activin signaling. Transgenic lines: cyp26a1, RA degrading enzyme ? inhibitor of RA signaling; dnfgfr1, dominant negative Fgf receptor 1 ? inhibitor of Fgf signaling; nog3, noggin3 (secreted Bmp antagonist) ? inhibitor of Bmp signaling. (I-O) axin2 expression is robustly detectable after inhibition of Activin (I), Igf (J), Hh (K), Notch (L), Fgf (M), RA (N) or Bmp (O) signaling for 6 hours using pharmacological compounds or heat shock-induced overexpression of pathway inhibitors. (P) axin2 expression is strongly reduced after inhibition of Wnt/&beta-catenin signaling for 6 hours using the pharmacological compound IWR-1 which stabilizes Axin. (A-P) Small arrowheads: amputation plane. Scale bars: whole mounts, 200 μm and 100 μm (C); sections, 100 μm. |
|
Interaction of Wnt/β-catenin signaling with other signaling pathways, related to Figure 6. (A) Overexpression of the Bmp inhibitor noggin3 (nog3) for 6 hours interferes with lef1 expression in hs:nog3 transgenic regenerates at 3 dpa. (B) ptc2 expression is detected in the basal epidermal layer and in (pre)osteoblasts (arrowhead). (C) dkk1 overexpression strongly interferes with regenerative growth which cannot be rescued by concomitant administration of RA via intraperitoneal injections. n = 5 fish, 6 rays each. Error bars indicate error of the mean. (D) fgf3 (blue) and mCherry (brown) transcripts co-localize in the distal-most blastema (arrowhead) in 7xTCF:mCherry transgenic regenerates. Note that samples were stained for mCherry expression only shortly, so that only the strongest expression domain in the distal blastema was detected. (A-D) Small arrowheads: amputation plane; scale bars: whole mounts, 200 μm and 100 μm (D); sections, 100 μm. |