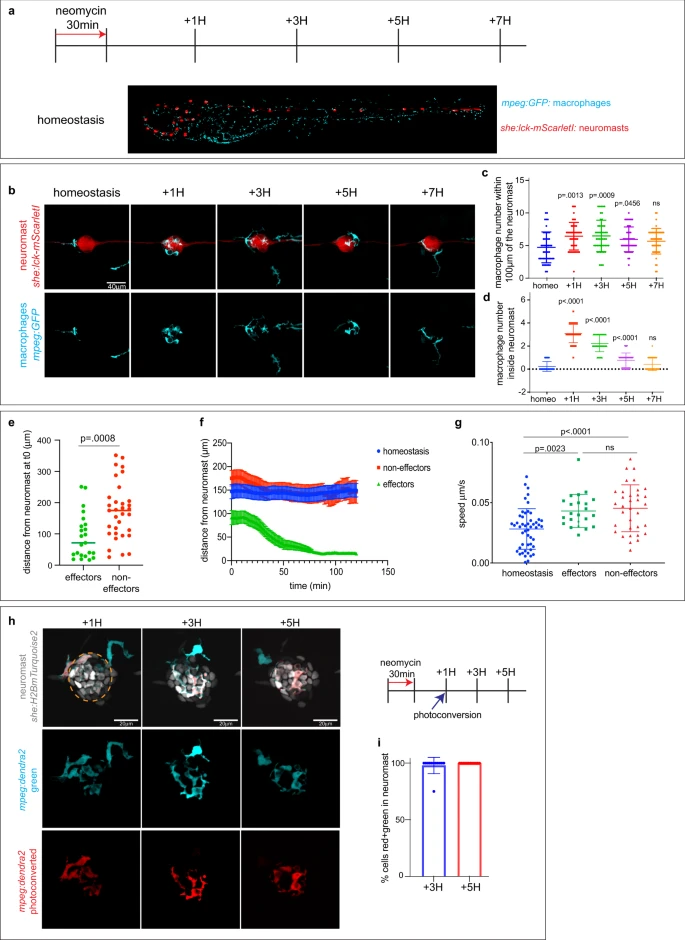
Fig. 1
a Schematics of neomycin regime and time point collections for the macrophage recruitment assay in the Tg(she:lck-mScarletI;mpeg:GFP) larvae. b Representative confocal images (projection of a 30 mm z-stack) for the macrophage recruitment assay. c, d Quantification of macrophages around (c) or inside (d) the neuromast. Each dot represents the number of macrophages per neuromast (3 neuromasts per larvae, 16 larvae per condition and 3 biological replicates). A 2-way ANOVA followed by a Tukey multiple comparison tests has been used to determine statistical significance. P-values represent a post-hoc (Tukey) test from each condition relative to homeostasis. e Quantification of the distance from the neuromast prior to HC death of effector and non-effector macrophages. Each dot represents a macrophage (8 larvae per condition and 3 biological replicates). P-value represents a two-tailed Student’s t-test. f Quantification of the distance from the neuromast over-time after HC death of effector and non-effector macrophages (8 larvae per condition and 3 biological replicates). g Quantification of the macrophage velocity of effectors and non-effectors macrophages. Each dot represents a macrophage (8 larvae per condition and 3 biological replicates). A 2-way ANOVA followed by a Tukey multiple comparison test has been used to determine statistical significance. P-values represent a post-hoc (Tukey) test between each condition. h Representative confocal images (projection of a 30 mm z-stack) for the macrophage photoconversion assay. Cells photoconverted (red) were inside the neuromast at the time of photoconversion while non-photoconverted cells (cyan) were not. Orange dotted line represents the photoconverted area. i Quantification of the ratio of photoconverted effector macrophages over time. Each dot represents the percentage of macrophages per neuromast (1 neuromast per larvae, 12 larvae per condition and 3 biological replicates). For all graphs, data are represented as mean ± SD except for (f) which is represented as mean ± SEM.