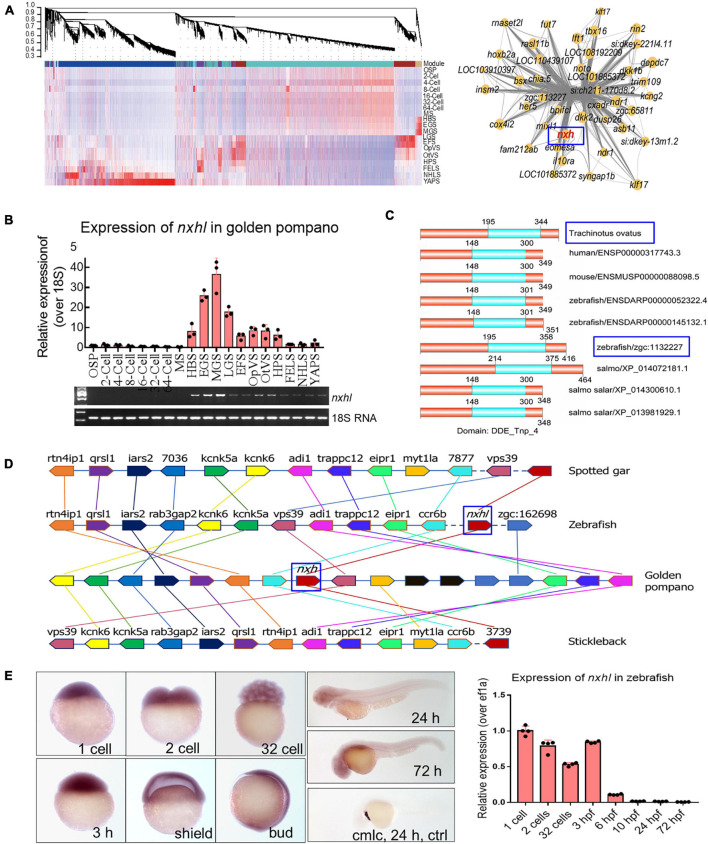
FIGURE 1
Evolutionary conservation of nxhl in vertebrate. (A) WGCNA analysis of embryonic development stages revealed gene-network modules enriched. HBS, EGS and MGS are classified into one group. OSP, oosperm; MS, morula stage; HBS, high blastula stage; EGS, early gastrula stage; MGS, middle gastrula stage; LGS, late gastrula stage; EFS, embryo formed stage; OpVS, optic vesicle stage; OtVS, otocyst vesicle stage; HPS, heart pulsation stage; FELS, formation of eye lens; NHLS, newly hatched larvae; YAPS, yolk absorption period. (B) Validation of expression level for nxhl by qPCR technology. 18s RNA was considered as internal marker. Gene structure of nxhl was showed at upper region. (C) Domains of nxhl and other homologous protein. The domain DDE_Tnp_4 was identified in SMART database (http://smart.embl.de/). (D) Micro-synteny analysis of nxh locus among spotted gar, zebrafish, gold pompano and stickleback. Two inversions and one insertion occurred in nxh locus region of golden pompano genomes. (E) The distribution of nxhl in zebrafish embryo by using whole-mount embryo in situ hybridization (WISH) and qPCR. Cmlc was used as positive control. Nxhl is mainly distributed in maternal developmental stages in zebrafish embryo.