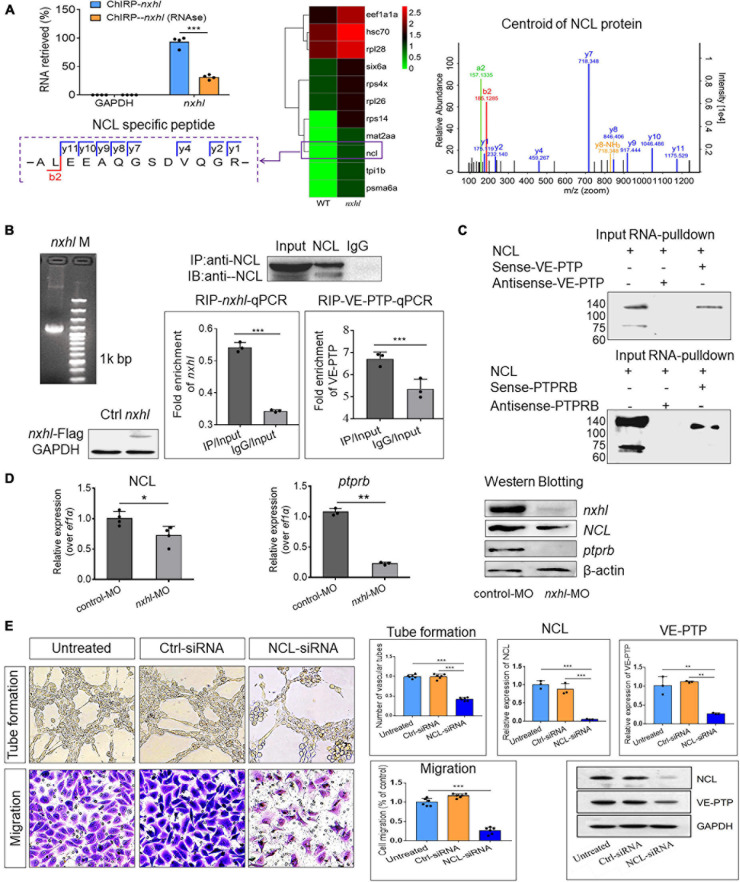
FIGURE 6
Nxhl regulates VE-PTP (ptprb) through interactions with NCL. (A) ChIRP-MS identification of nxhl RNA binding proteins using zebrafish tissues. qPCR identification of nxhl RNA in the eluted RNAs. Graph shows more than 90% nxhl RNA was retrieved, and no GAPDH was detected. Heat map shows major proteins are enriched and significantly (change fold > 2 and p < 0.05) retrieved by nxhl and control probes, analyzed by LC/MS-MS. NCL protein (purple boxed) was selected as candidate for follow-up study. The Centroid of NCL protein shows that NCL protein is pulled down and identified by LC/MS-MS. The specific peptide identifies NCL protein. (B) The interaction between nxhl, VE-PTP mRNA and NCL protein by RIP-qPCR assay. The mRNA expression of nxhl was determined by qPCR and Western blotting against Flag antibody and used to identify the successful expression of pcDNA3.1- Flag-nxhl plasmid in 293T cells. Bars show the interaction between nxhl mRNA and NCL protein. The interaction between VE-PTP mRNA and NCL protein is shown too, and qPCR shows the detection for VE-PTP mRNA expression in the NCL-pulled down RNA. (C) The interaction between nxhl, VE-PTP mRNA and NCL protein by pull down assay. Gels show the interaction between VE-PTP mRNA and NCL protein. Western blotting was performed to detect NCL protein in the VE-PTP-biotin probe -pulled down proteins in 293T cells. The interaction between ptprb mRNA and NCL protein is shown too. (D) Loss of nxhl affects the expression of NCL at both mRNA and protein levels. The mRNA expression of NCL and ptprb were determined by qPCR. The total NCL protein, total nxhl and ptprb protein in zebrafish tissues from knock-down group and control were detected by Western blotting using specific antibodies (details see Materials and methods). β-actin was used as internal control. (E) Silence of NCL inhibits angiogenesis and expression of VE-PTP. The tube formation and cell migration potential of HUVECs treated with NCL-siRNA was determined by using transwell chambers as described in the “Materials and methods” section. Scale bars, 20 μm. Representative images of cells stained in NCL-siRNA treated HUVEC cells. The expression of NCL and VE-PTP was quantified by qPCR. Protein levels of NCL and VE-PTP were examined by using Western blotting post silence of NCL. GAPDH antibody was used as internal control. The data represent as mean ± SEM from three independent experiments. *p < 0.05, **p < 0.001, ***p < 0.0001 represents statistically significant.