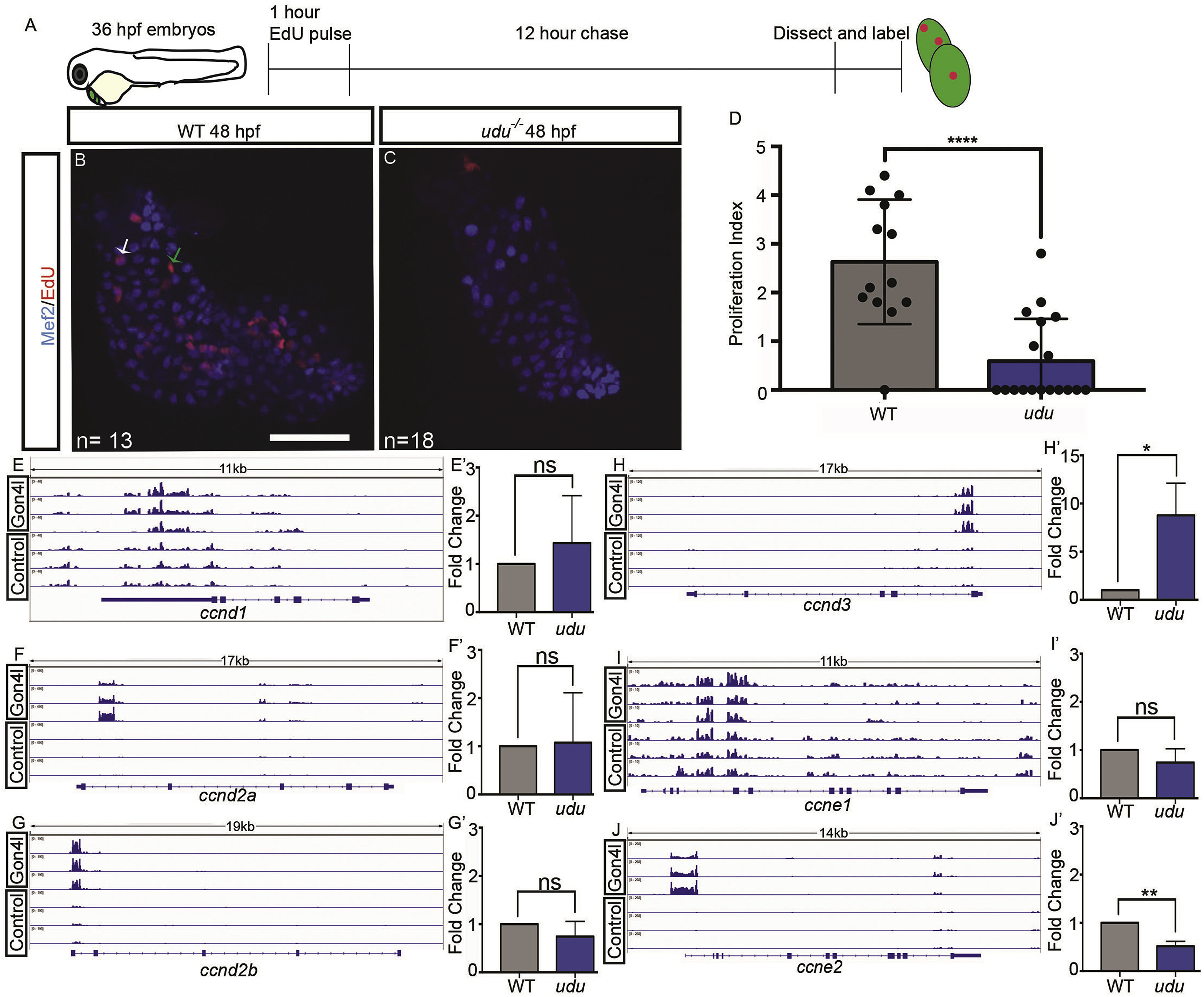
Fig. 3 EdU incorporation is reduced in the hearts of udu−/− embryos. Experimental workflow for EdU labeling (A). EdU labeling (red) in 48 hpf WT hearts (B) and udu−/− hearts (C) with cardiomyocytes labeled by Mef2 (blue). Arrows showing EdU + cardiomyocyte (white) and non-cardiomyocyte (green). Graph showing the proliferative index for EdU in WT (grey) and udu−/−(blue) cardiomyocytes (D). qRT-PCR for ccnd1 (E), ccnd2a (F), ccnd2b (G), ccnd3 (H), ccne1 (I), and ccne2 (J) performed on RNA extracted from hearts isolated from 36 hpf WT (grey) and udu−/−(blue) embryos. Results are shown as fold change of gene expression in udu−/− embryos compared to WT and results were standardized to gapdh expression. DamID-seq Genome browser tracks of Gon4l-Dam and GFP Control-Dam at ccnd1 (E′), ccnd2a (F′), ccnd2b (G′), ccnd3 (H′), ccne1 (I′), and ccne2 (J′) loci. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, error bars = SEM. Scale bar represents 50 μm.
Reprinted from Developmental Biology, 462(2), Budine, T.E., de Sena-Tomás, C., Williams, M.L.K., Sepich, D.S., Targoff, K.L., Solnica-Kreze, L., Gon4l/Udu Regulates Cardiomyocyte Proliferation and Maintenance of Ventricular Chamber Identity During Zebrafish Development, 223-234, Copyright (2020) with permission from Elsevier. Full text @ Dev. Biol.