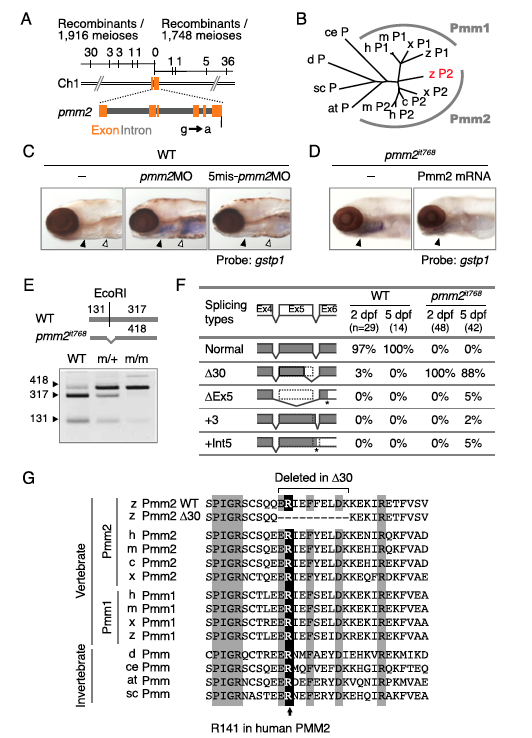
Fig. S2
The gene responsible for the it768 mutant is pmm2. (A) A physical map of the it768 locus on the zebrafish chromosome 1. The numbers show recombinants identified by a simple sequence-length polymorphism analysis using polymorphic markers. The G-to-A mutation at position 1 of the 5′ splice site in the intron 5 was found in the mutant allele. (B) A phylogenetic tree constructed by the neighbor-joining method for the full-length amino acid sequences of Pmm proteins. (C) gstp1 expression at 5 dpf in WT or pmm2it768 larvae injected or not with 1 pmol each of the indicated morpholino oligonucleotides. The closed and open arrowheads indicate the gills and liver, respectively. (D) gstp1 expression at 5 dpf in WT or pmm2it768 larvae injected or not with mRNA encoding zebrafish Pmm2 protein. (E) Results of the RT-PCR analysis using total RNA isolated from WT, pmm2it768/+ (m/+), or pmm2it768/it768 (m/m) larvae at 5 dpf and primers designed to amplify partial cDNA corresponding to the exon 5–intron 5–exon 6 region. EcoRI cut the cDNA from the normally spliced mRNA. (F) Results of the cDNA analysis prepared from total RNA of whole bodies in WT or pmm2it768 larvae at the indicated dpf. The numbers of the sequenced cDNA and the percentages of the indicated splicing types are shown. The asterisks indicate stop codons. (G) Amino acid sequences corresponding to exons 5 and 6 of the vertebrate and invertebrate Pmm proteins. The highlighted amino acid residues are highly conserved among eukaryotes. at, Arabidopsis thaliana; c, chicken; ce, Caenorhabditis elegans; d, Drosophila melanogaster; h, human; m, mouse; sc, Saccharomyces cerevisiae; x, Xenopus laevis; z, zebrafish.