Fig. 3
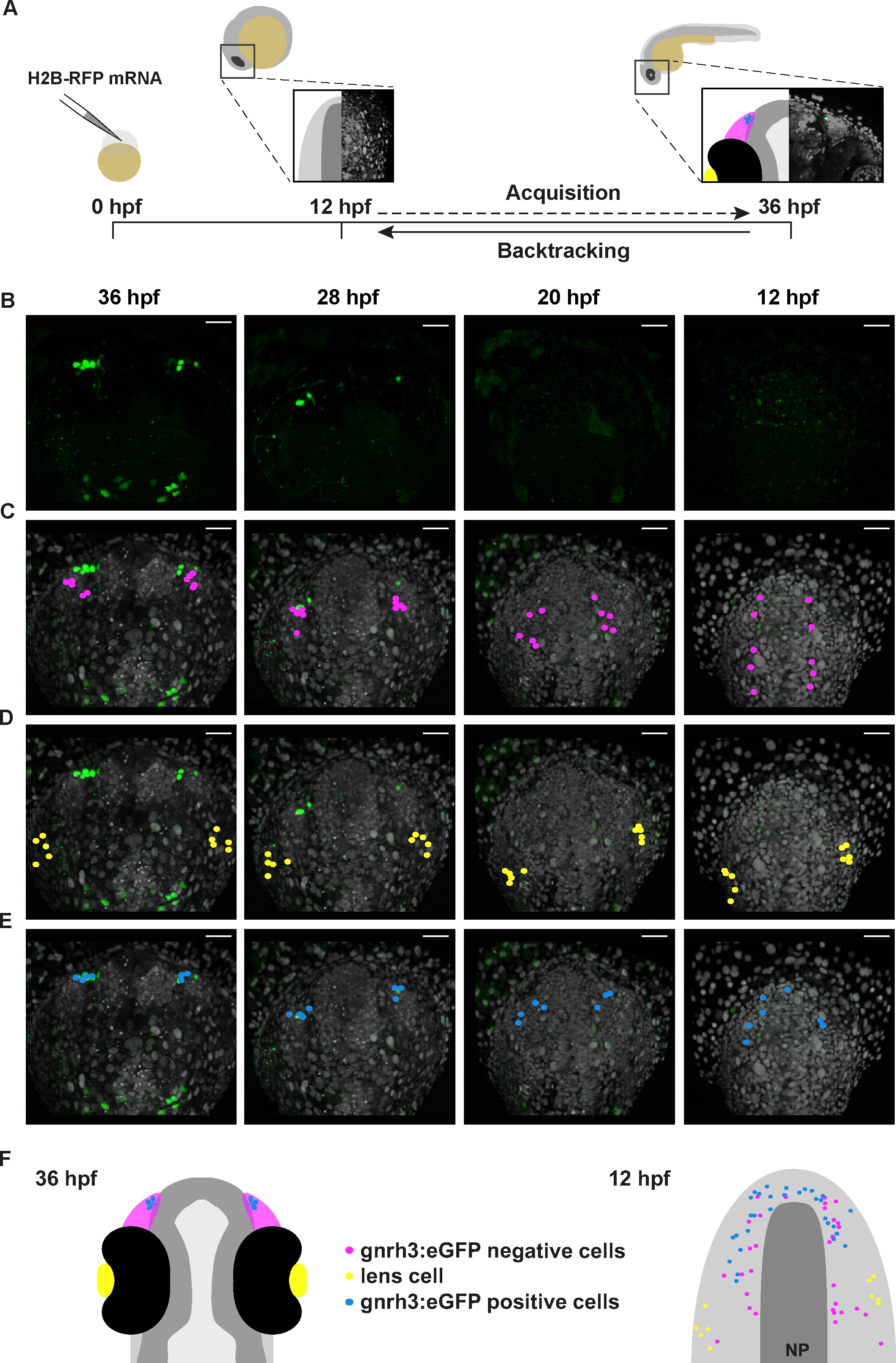
Lineage reconstruction reveals an anterior preplacodal ectoderm origin for Gnrh3 neurons: backtracking.
(A) Schematic representation of the backtracking strategy. Synthetic mRNAs encoding Histone2B-RFP (H2B-RFP) were injected into Tg(gnrh3:eGFP) transgenic embryos, which were subsequently imaged from 12 to 36 hpf. The lineages of various populations of cells were manually retraced by backtracking H2B-RFP+ nuclei to their position at the beginning of the time-lapse series. (B–E) Confocal projections from a representative 4D dataset at 36, 28, 20 and 12 hpf showing the GFP channel alone (B), the position of the nuclei backtracked from 10 gnrh3:eGFP-negative (C, pink), 10 lens (D, yellow) and 7 gnrh3:eGFP-positive (E, blue) cells at each timepoint. The embryo is shown with anterior up; GFP expression detected caudally is ectopic and does not reflect endogenous gnrh3 expression. Scalebars represent 40 μm. (F) Schematic representation of an embryonic head at 36 hpf and the anterior neural plate (NP, dark grey) and adjacent preplacodal ectoderm (light grey) at 12 hpf. The 36 hpf head shows the colour code of the backtracked lineages, and the position of backtracked nuclei at 12 hpf is indicated in the preplacodal ectoderm. The 12 hpf representation shows the results obtained from 9 epithelia (5 embryos) for 30 gnrh3:eGFP-positive cells and 32 gnrh3:eGFP-negative cells; the 10 lens cells were backtracked from a pair of epithelia in a single 4D dataset only.