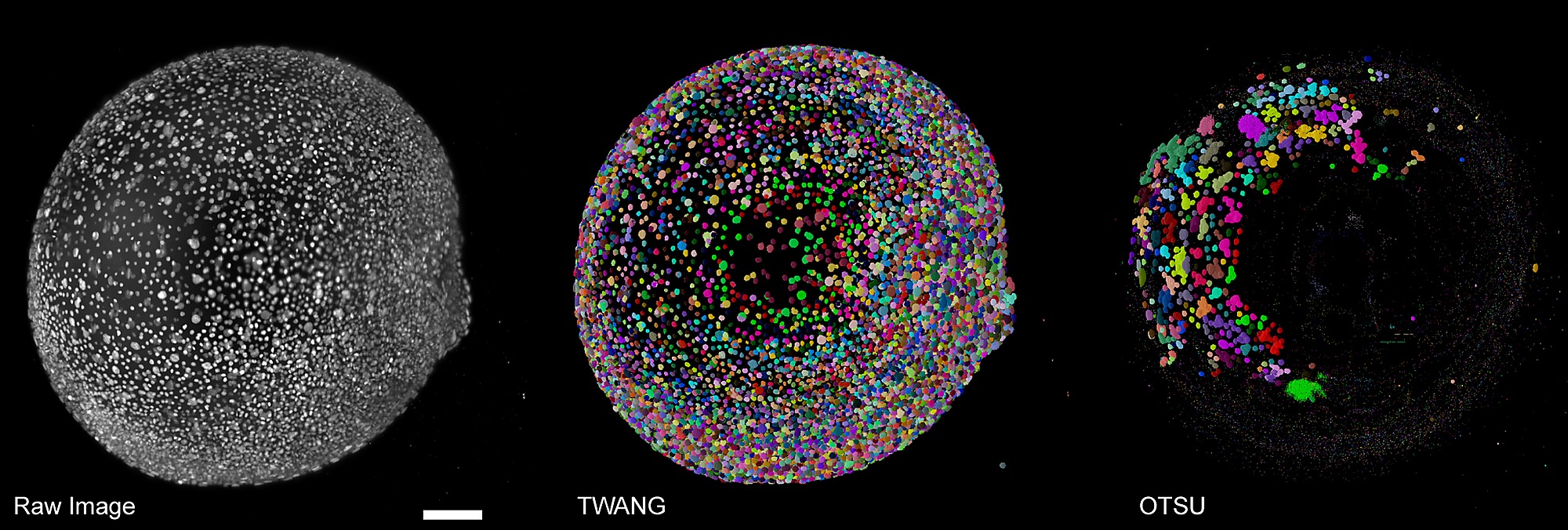
Fig. s8
Exemplary comparison of applying OTSU and TWANG to a large-scale light-sheet microscopy image of fluorescently labeled cell nuclei of a zebrafish embryo(single time point at about 9 hours post fetilization).
OTSU completely failed to extract meaningful information in this case and only captured a few correct nuclei by chance. TWANG could resolve most of the nuclei correctly and by design largely avoids under-segmentation errors. As the results provided by the plain OTSU algorithm were not properly extracting the embryonic shape, the watershed-based post-processing steps used in OTSUWW and OTSUWW+U were skipped. The results of TWANG+U are visually indiscernible from the plain TWANG results and we thus only show one panel for convenience. Furthermore, due to the image size of about 5 GB per frame, applying the 3D watershed algorithm on the entire image was not possible given the memory limitations of 32 GB on our test computer. A lack of ground truth for these large-scale data sets only allowed a qualitative comparison (see [39] for details). Scale bar: 100μm.