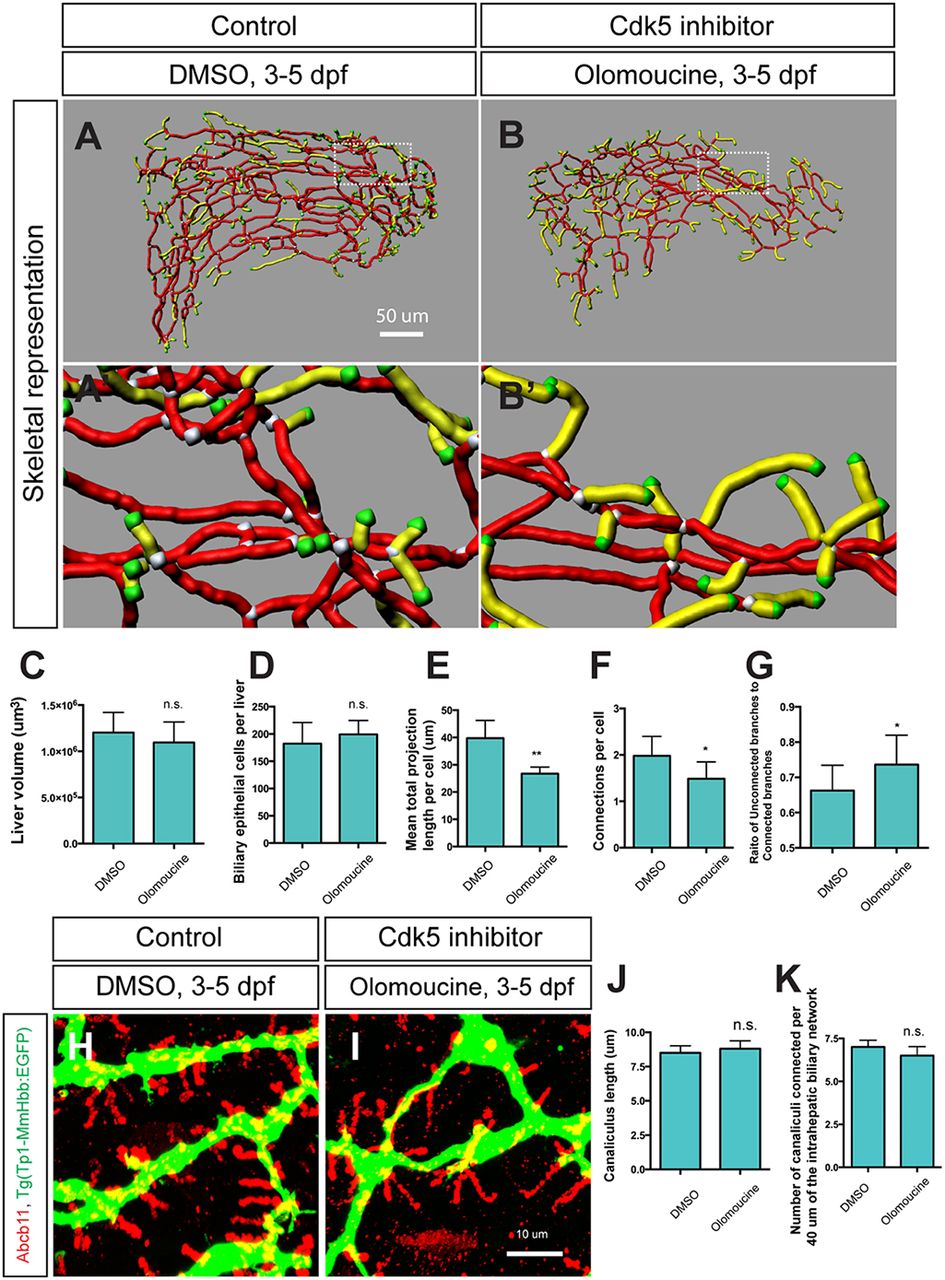
Fig. 2
Cdk5 regulates intrahepatic biliary network formation. (A,B) Skeletal representation of the intrahepatic biliary network in Tg(Tp1-MmHbb:EGFP)um14 larvae treated from 3 to 5 dpf with DMSO (A) or the Cdk5 inhibitor olomoucine (B) computed based on Tg(Tp1-MmHbb:EGFP)um14 expression at 5 dpf. Boxed regions in A and B are magnified in A′ and B′. Nodes, end points, connected (node-node) and unconnected (node-end point) branches are colored as in Fig. 1F. (C-G) Measurements of intrahepatic biliary network structures by the skeletal analysis algorithm of the DMSO-treated and olomoucine-treated Tg(Tp1-MmHbb:EGFP)um14 larvae at 5 dpf. (C) Mean liver volume at 5 dpf. Liver volume was not affected by olomoucine treatment. (D) Average number of Tg(Tp1-MmHbb:EGFP)um14-positive biliary epithelial cells in the liver at 5 dpf. (E) Mean total projection length of the intrahepatic biliary network normalized to biliary epithelial cell number. (F) Number of connected branches normalized to biliary epithelial cell number. (G) Ratio of connected to unconnected branches. n=5 for all measurements. (H,I) Projected images of confocal z-stacks of the liver in DMSO-treated (H) or olomoucine-treated (I) larvae visualized for the bile canaliculus marker Abcb11 (red) and Tg(Tp1-MmHbb:EGFP)um14 (green) expression at 5 dpf. (J) Average length of canaliculus measured based on Abcb11 expression. In total, 135 canaliculi were analyzed. (K) The number of canaliculi connected per 40 μm of the Tg(Tp1-MmHbb:EGFP)um14-expressing intrahepatic biliary network (n=10 for each condition). Error bars are s.d. *P<0.05, **P<0.01; n.s., not significant.