Fig. 1
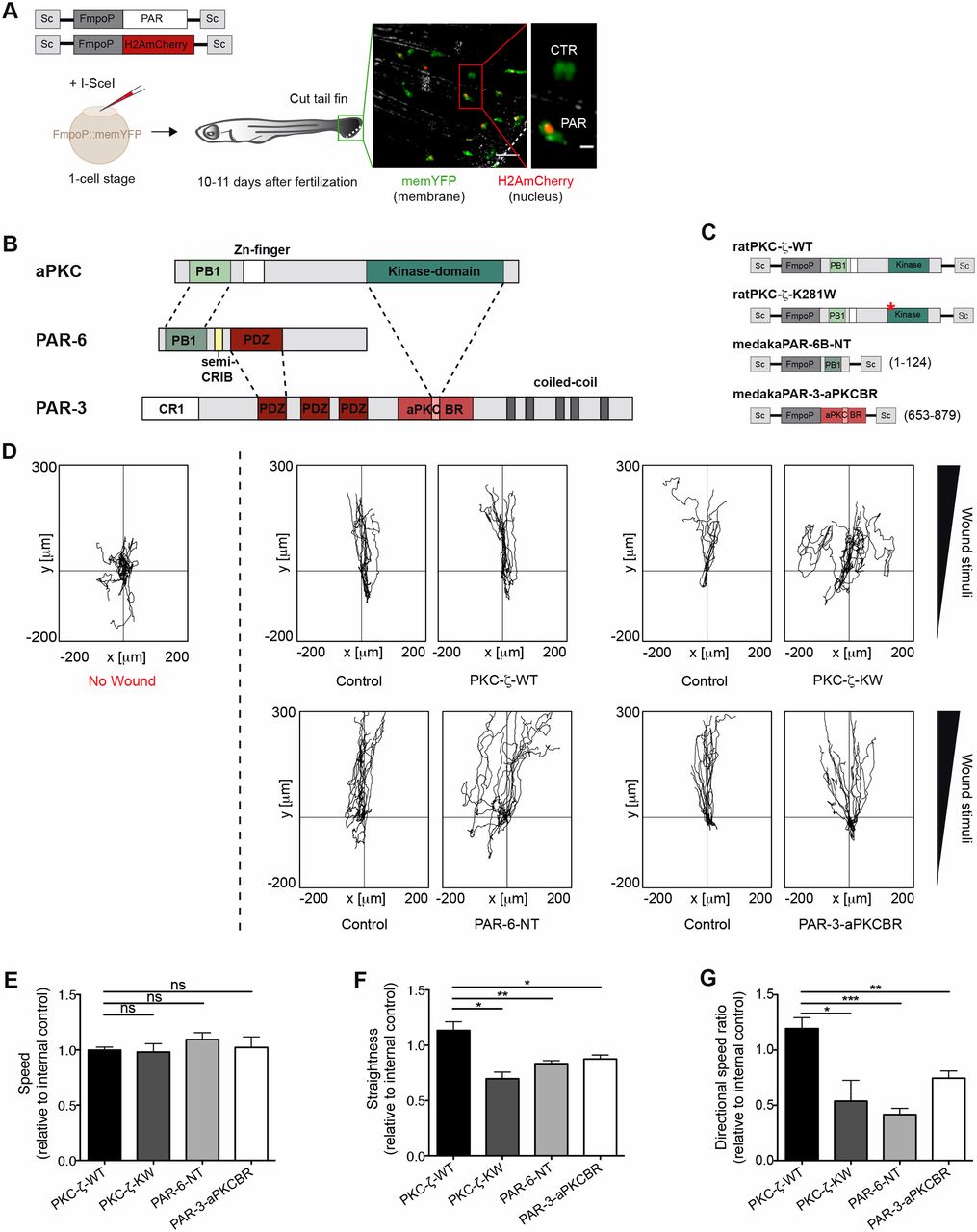
The PAR complex promotes wound-directed migration of myeloid cells in vivo. (A) TG(FmpoP::memYFP) embryos were injected at the one-cell stage with a 1∶1 mixture of DNA coding for H2AmCherry as a nuclear reporter and one of the PAR transgenes, driven by the myeloid-specific Fmpo promoter and flanked by I-SceI integration sites (Sc), in the presence of I-SceI meganuclease. Wounded larvae with mosaic expression of H2AmCherry in tailfin myeloid cells were imaged. The dashed line represents the wound. The inset shows a transgenic cell (Cherry+; PAR) and the endogenous control (Cherry−; CTR). Scale bars: 50 µm (left panel); 10 µm (inset). (B) Domains of interaction between members of the mammalian PAR complex. Connecting lines indicate regions of the proteins that interact with one another. PB1, phagocyte oxidase/Bem1 domain; Zn, Zinc finger motif; Kinase, catalytic domain; CRIB, Cdc42/Rac interactive binding motif; PDZ, PSD-95/Dlg/Zona occludens-1 domain; CR1, conserved region 1; aPKCBR, aPKC-binding region. A predicted coiled-coil region is also shown. (C) Schematics of the constructs used to perturb the function of the PAR complex in myeloid cells. Numbers refer to amino acid positions. *K to W mutation at codon 281. NT, N-terminal domain. (D) 2D tracks of individual leukocytes migrating in the tailfin of unwounded fish (left panel) or towards the tailfin wound (right panels). No wound, n = 11; control/PKC-ζ-WT, n = 12; control/PKC-ζ-KW, n = 10; control/PAR-6-NT, n = 13; control/PAR-3-aPKCBR, n = 13). Tracks are from one representative experiment of at least three independent experiments. (E–G) Quantification of 2D (E) speed, (F) path straightness and (G) directional speed ratio of myeloid cells during the wound response. Data are expressed as the mean±s.e.m. of at least three separate experiments (PKC-ζ-WT, n = 27 cells in three larvae; PKC-ζ-KW, n = 27 cells in three larvae; PAR-6-NT, n = 45 cells in four larvae; PAR-3-aPKCBR, n = 64 cells in five larvae); *P<0.05; **P<0.01; ***P<0.001; ns, non-significant (two-tailed unpaired Student's t-test). See also supplementary material Fig. S1; Movies 1, 2.