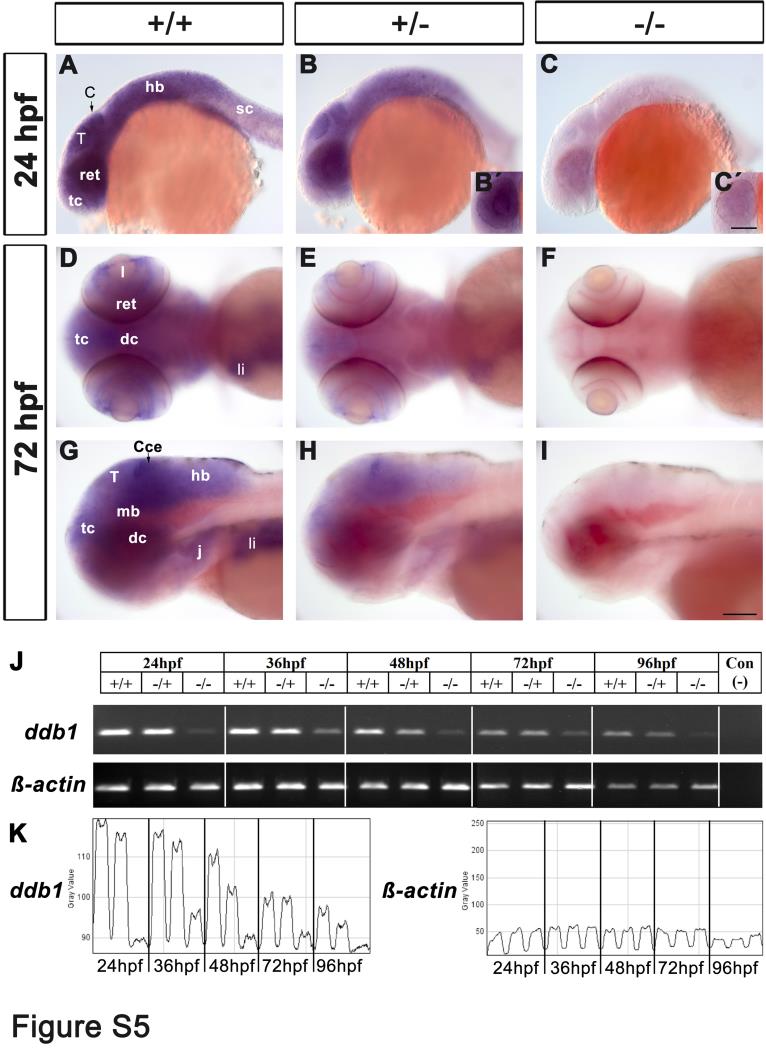
Fig. S5
Reduction of ddb1 mRNA in homozygous ddb1m863 mutants by WISH and RT-PCR.
(A-I) WISH analysis of ddb1 transcripts revealed reduced expression in heterozygous (B, E, H) and near-absent expression in homozygous ddb1m863 mutants (C, F, I) compared to wild type siblings (A, D, G) at 24 hpf (A-C) and 72 hpf (D-I). Planes focus on the retina of heterozygous embryos (B′) and homozygous mutants (C′). Lateral views (A-C; G-I) and dorsal views (D-F). Abbreviations used: Cce, cerebellum; dc, diencephalon; hb, hindbrain; j, jaw; I, lens; li, liver; mb, midbrain; ret, retina; sc, spinal cord; T, tectum; tc, telencephalon. Anterior is towards the left. Scale bars in C′ for B′-C′, in I for others: 100 µm. (J) RT-PCR analysis of ddb1 transcript levels during development stages from 24 hpf to 96 hpf in wild type, heterozygous and homozygous m863 mutants. β-actin was used as an internal control, and the negative control was template-free. (K) The RT-PCR gel was quantified densitometrically and traces of ddb1 PCR signal and of internal β-actin control were shown together with the wild type, heterozygous and homozygous m863 mutant samples adjacent to each other within each frame showing one developmental stage. Transcription of ddb1 was progressively downregulated in wild type embryos along the development. Compared to wild type siblings, the transcription level of ddb1 was slightly decreased in heterozygous embryos/larvae, but strongly reduced in homozygous ddb1m863 mutants.