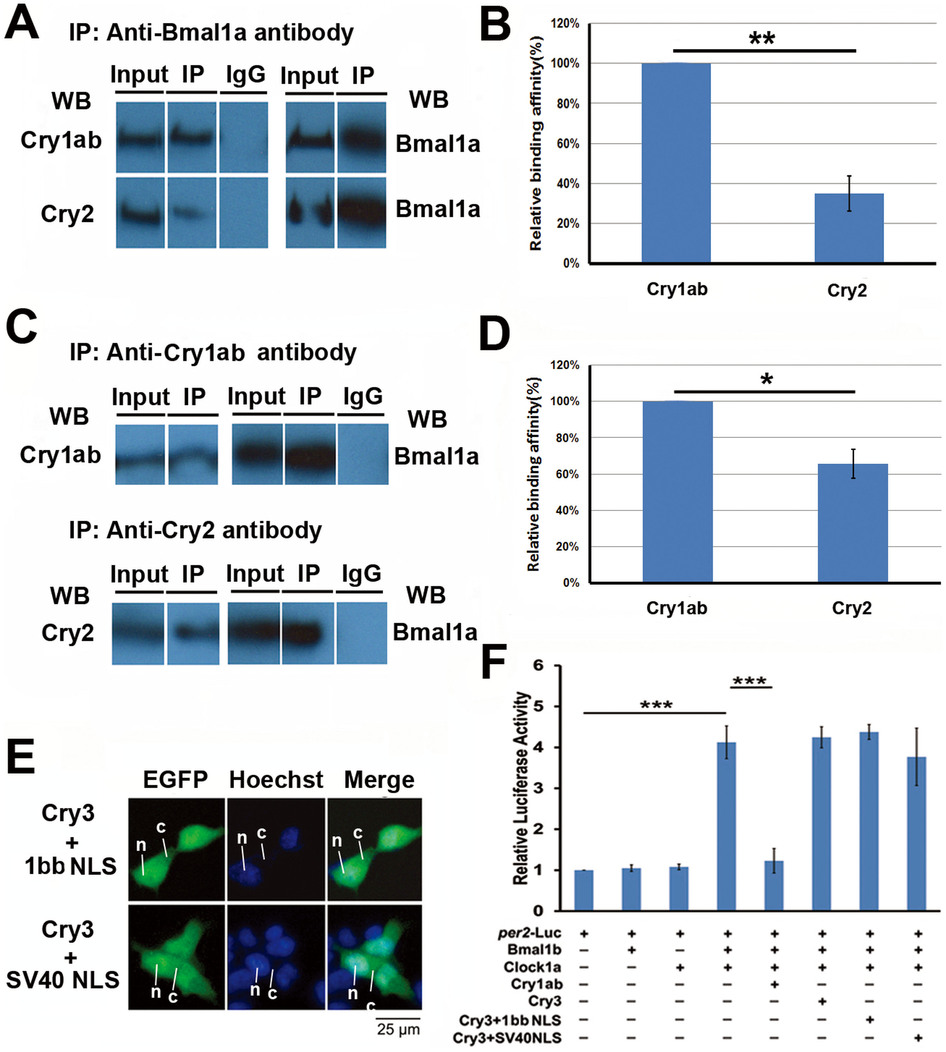
Fig. 8
Mechanisms for non-repression of zebrafish Cry2 and Cry3 on Clock:Bmal mediated transcription.
(A) Co-immunoprecipitation (Co-IP) experiments. Zebrafish cry1ab and cry2 constructs were co-transfected into HEK293 cells with per2-Luc reporter and bmal1a, and clock1a, respectively. Then the anti-Bmal1a polyclonal antibody was used to pull down proteins bound to Bmal1a, which was identified by the anti-Cry1ab and anti-Cry2 polyclonal antibodies, respectively. (B) The relative protein binding affinity of Cry1ab and Cry2 to Bmal1a-Clock1a complex. The relative protein binding affinity was quantified with Image pro-Plus6.0. Each value is the mean ± SD of three independent experiments. Results were analyzed by Student′s t-test. Horizontal lines indicate columns compared for significance. One star on the line shows 0.01 < p < 0.05. (C) Protein binding affinity comparison of Cry1ab and Cry2 to Bmal1a:Clock1a complex. Zebrafish cry1ab and cry2 constructs were co-transfected into HEK293 cells with per2-luc reporter and Bmal1a-Clock1a combination, respectively. Then anti-Cry1ab or anti-Cry2 polyclonal antibody was used to collect the proteins bound to Bmal1a, which was identified by the anti-Bmal1a polyclonal antibody. (D) The relative protein binding affinity of Cry1ab and Cry2 to the Clock1a-Bmal1a complex from Fig 8C. The relative protein binding affinity was quantified by Image pro-Plus6.0. Each value is the mean ± SD of three independent experiments. Results were analyzed by Student′s t-test. Horizontal lines indicate columns compared for significance. Two stars on the line means p < 0.01, (E) Sub-cellular localization of zebrafish Cry3 fused to either Cry1bb NLS from Fig 7A or a SV40 NLS. The Cytoplasmic-nuclear distribution of these two DNA constructs was detected by GFP signals (green) and the nucleus identified by Hoechst (blue). The leader lines and letters N, C indicate the location of nucleus and cytoplasm, respectively. (F) Inhibitory activities of zebrafish Cry3 fused to either Cry1bb NLS or a SV40 NLS determined by dual Luciferase reporter assays. The two DNA constructs were co-transfected with per2-Luc reporter and a Clock1a-Bmal1b combination, respectively. A Renilla Luciferase was added in each transfection to normalize transfection efficiency. The figure shows the mean and SD (error bar) of the two independent experiments (triplicate for each experiment). Results were analyzed by ANOVA.