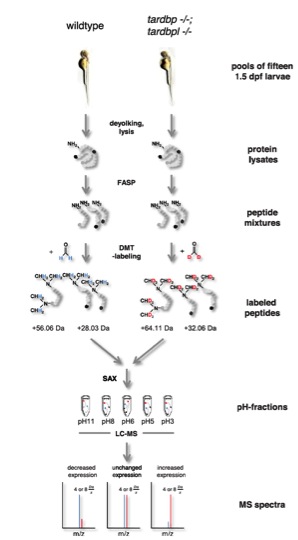
Fig. S7 Schematic view of the quantitative proteomic analysis. Pools of 16 1.5-dpf embryos were deyolked and lysed consecutively. Protein lysates were tryptically digested using the filter-assisted sample preparation technique (FASP). Tryptic peptides ending either on arginine (depicted in black) or a lysine (depicted in dark gray) were labeled using differential stable isotope dimethyl labeling (DMT-labeling). Peptides gained from wild-type embryos were labeled “light” using undeuterated formaldehyde, whereas peptides gained from tardbp-/-;tardbpl-/- mutant embryos were labeled “heavy” using deuterated formaldehyde. Dimethylation occurs at free amine-groups resulting in the addition of four methyl-groups to lysine-containing tryptic peptides whereas only two methyl-groups are added to arginine-containing tryptic peptides; this results in a shift of +56.06 Da or +28.03 Da, respectively, for the light-labeledpeptides. Heavy-labeled peptides are shifted by further +8 Da or +4 Da, respectively, because of the two deuterium atoms contained in every methyl-group. After labeling, peptides from both experimental groups were combined and fractionated using stage-tip-based anion-exchange chromatography (SAX). Mass spectrometry was done for every of the five pH-fractions in two technical replicates. Abundance levels of peptides can be calculated from the intensity ratio between the isotope clusters resulting from the light and heavy peptides.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Proc. Natl. Acad. Sci. USA