Fig. S2
zCxcl8 Imposes an Orthotactic Bias on Neutrophil Random Walk (Related to Figure 2)
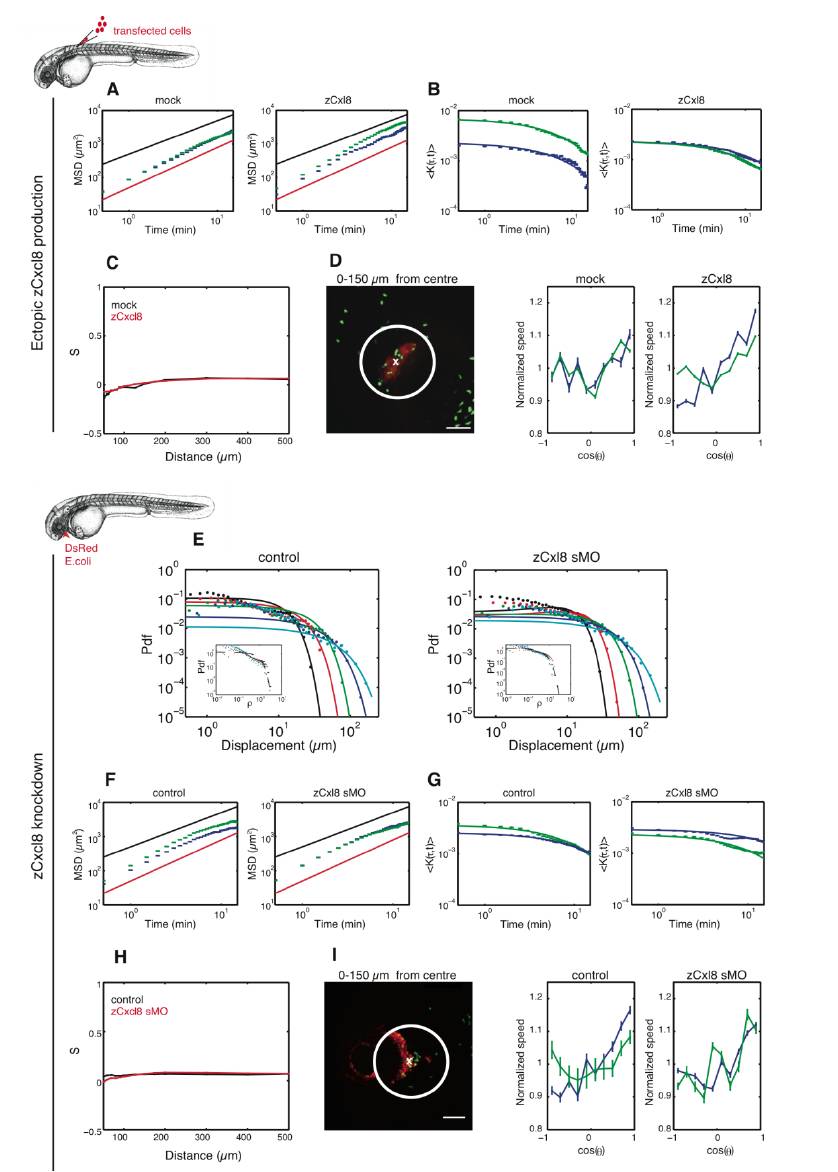
(A) Evolution of the Mean Square Displacement (MSD) with time in log-log scale. Blue color is associated to cells inside a radius of 100μm and green color is associated to the complete set of cells in the image field. Red line slope is 1.2 and black line slope is 1. Note that the MSD starts super diffusive with a slope (log-log scale) close to 1.2 but eventually returns to a slope close to 1 in both cases.
(B) Evolution of the normalized displacement correlation K(τ,t) with time τ. Color code is identical to A. Solid lines are exponential fits.
(C) Evolution of the nematic order parameter S with the distance from the center of the transplant.
(D) (Left) Representative reference image showing the mCherry/zCxcl8-expressing transplanted cells in red, GFP+ neutrophils in green and a 150μm radius in white. (Right) Evolution of the average normalized speed with the orientation of the motion. Color code is identical to A. Note the strong discrepancy between the green and blue curve in the zCxcl8 case, which is not observed in the control case.
(E) Evolution of the propagator, i.e. the probability density function (Pdf) of neutrophil displacement r(t) at different times t, in larvae treated or not with zCxcl8 sMO. Colors are associated with times: black for t=1 min, red for t=2 min, green for t=4 min, blue for t=8 min and cyan for t=16 min. Plots are in log-log scale. Inserts are the collapsed curves, P(ρ), into a unique Gaussian. Black line is the Gaussian fit.
(F) Evolution of the Mean Square Displacement (MSD) with time in log-log scale. Blue color is associated to cells inside a radius of 100μm and green color is associated to the complete set of cells. Red line slope is 1.2 and black line slope is 1. Note that the MSD starts super diffusive with a slope (log-log scale) close to 1.2 but returns to a slope close to 1 in both cases.
(G) Evolution of the normalized displacement correlation K(τ,t) with time τ.. Color code is identical to A. Solid lines are exponential fits.
(H) Evolution of the nematic order parameter S with the distance from the center of the infection site.
(I) (Left) Representative image showing DsRed+ E. coli in red, GFP+ neutrophils in green and a 150μm radius in white. (Right) Evolution of the average normalized speed with the orientation of the motion. Color code is identical to A. Note the strong discrepancy between the green and blue curve in the control case, which is less marked in the case of the zCxcl8 knockdown.
(A-I) Data are from 4 larvae in 2-4 experiments. (A, B, C, D, F, G, H, I) Error bars, SEM
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Curr. Biol.