Fig. S1
Cloning and expression pattern analyses of the crb2a and crb2b genes in developing zebrafish retina, related to Figure 2.
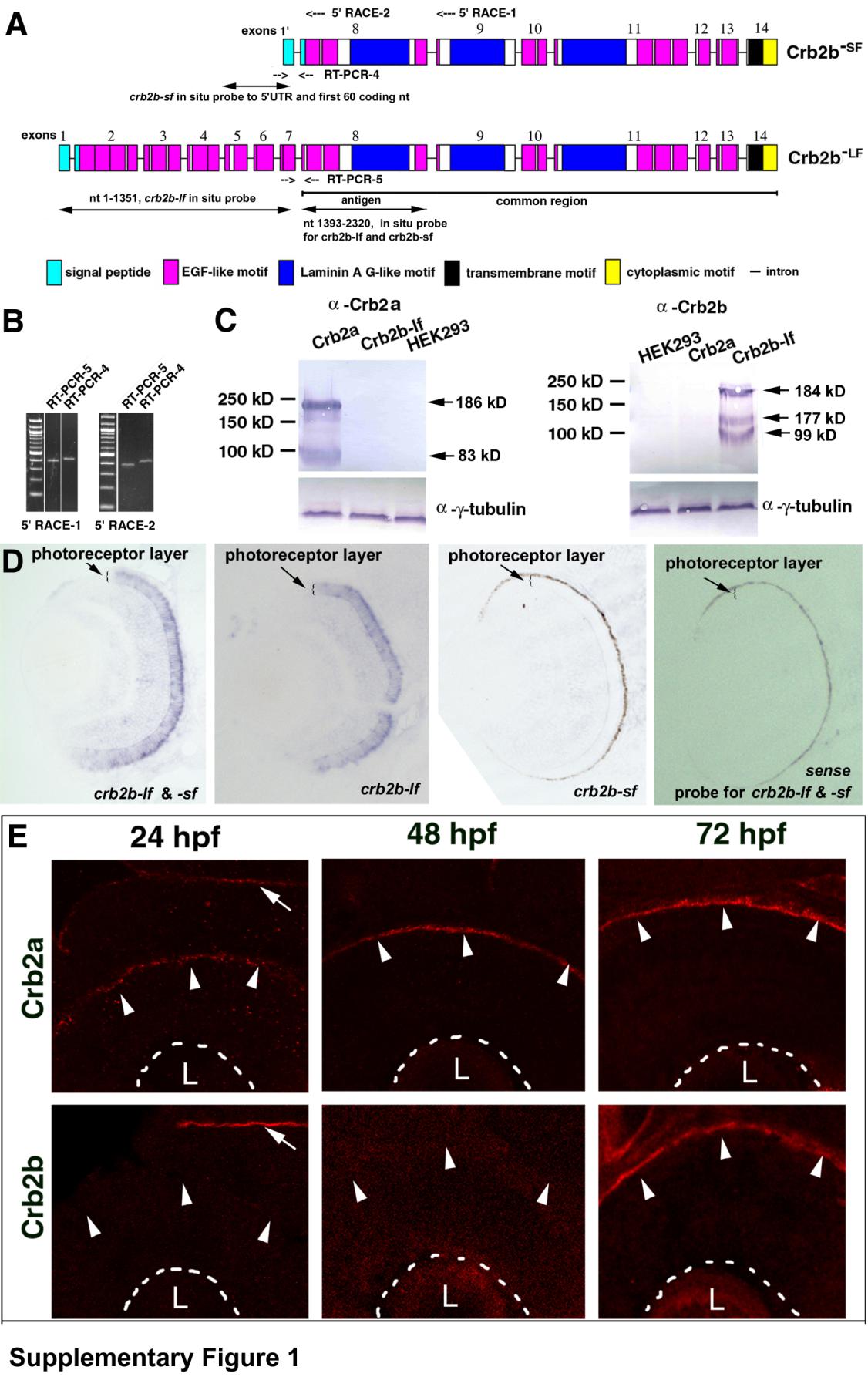
A. A schematic shows the exon-intron structural differences between crb2b-lf and crb2b-sf (not to scale). The exons are numbered from the 5′ end to the 3′ end. Primer binding sites for two 5′RACE reactions are indicated at the top of the chart. The regions amplified by RT-PCR reactions 4 and 5 are indicated with opposing arrows. The common region of the crb2b-lf and crb2b-sf transcripts is underlined. The antigenic region for making anti-Crb2b-lf amino acids 466-773 antibodies and the regions that the in situ probes bind to are indicated with double-headed arrows. B. The similar expression levels of crb2b-sf and crb2b-lf was confirmed by RT-PCR reactions 4 and 5, which used two different sets of cDNA prepared from 5′RACE-1 and 5′RACE-2. C. The specificity of the anti-Crb2a and anti-Crb2b antibodies was verified by Western blotting analyses of the lysates of Crb2a-, and Crb2b-lf-expressing HEK293 cells. The predicted molecular weights of Crb2a and Crb2b-lf are 161 and 160 kD, respectively. Western blots revealed higher apparent molecular weights for both Crb2a and Crb2b-lf, suggesting the presence of post-translational modifications. In addition, smaller-than-expected bands were detected, likely due to degradation or early termination of translation. The anti--tubulin blots served as sample loading controls. The specificity of the antibodies was also confirmed by the differences in their immnostaining patterns (Figures S1E and 2), as well as by the lack of immunoreactivity of crb2am289 mutant retinas to the anti-Crb2a antibodies (Figure 5B). The current data indicate that our previous immunohistological analyses of Crb expression patterns in zebrafish retina with the antibodies against the intracellular domains of mouse CRB1 and CRB2 were not completely accurate because both antibodies should have cross-reacted with zebrafish Crb2a and Crb2b due to high sequence similarity (Wei et al., 2006a). D. In situ hybridization analysis revealed that Crb2b expression is restricted in the photoreceptor layer. However, the probe designed to recognize crb2b-sf did not work, presumably because the probe is too short (246 nt). As expected, the negative control sense probe did not stain the retina. E. Crb2a is expressed at the apical surface (arrowheads) of the retina throughout retinal development, whereas Crb2b expression was detectable at the apical retinal surface at 72 hpf after embryonic photoreceptor differentiation but not at 24 and 48 hpf (arrowheads). The basal surfaces of the retina are marked by dashed lines. The arrows indicate the expression of Crb2a and Crb2b at the apical surface of the brain neuroepithelium, indicating that the lack of Crb2b signals in 24-hpf and 48-hpf retinas was not due to technical errors. The lenses are indicted with letters L.
Reprinted from Developmental Cell, 22(6), Zou, J., Wang, X., and Wei, X., Crb Apical Polarity Proteins Maintain Zebrafish Retinal Cone Mosaics via Intercellular Binding of Their Extracellular Domains, 1261-1274, Copyright (2012) with permission from Elsevier. Full text @ Dev. Cell