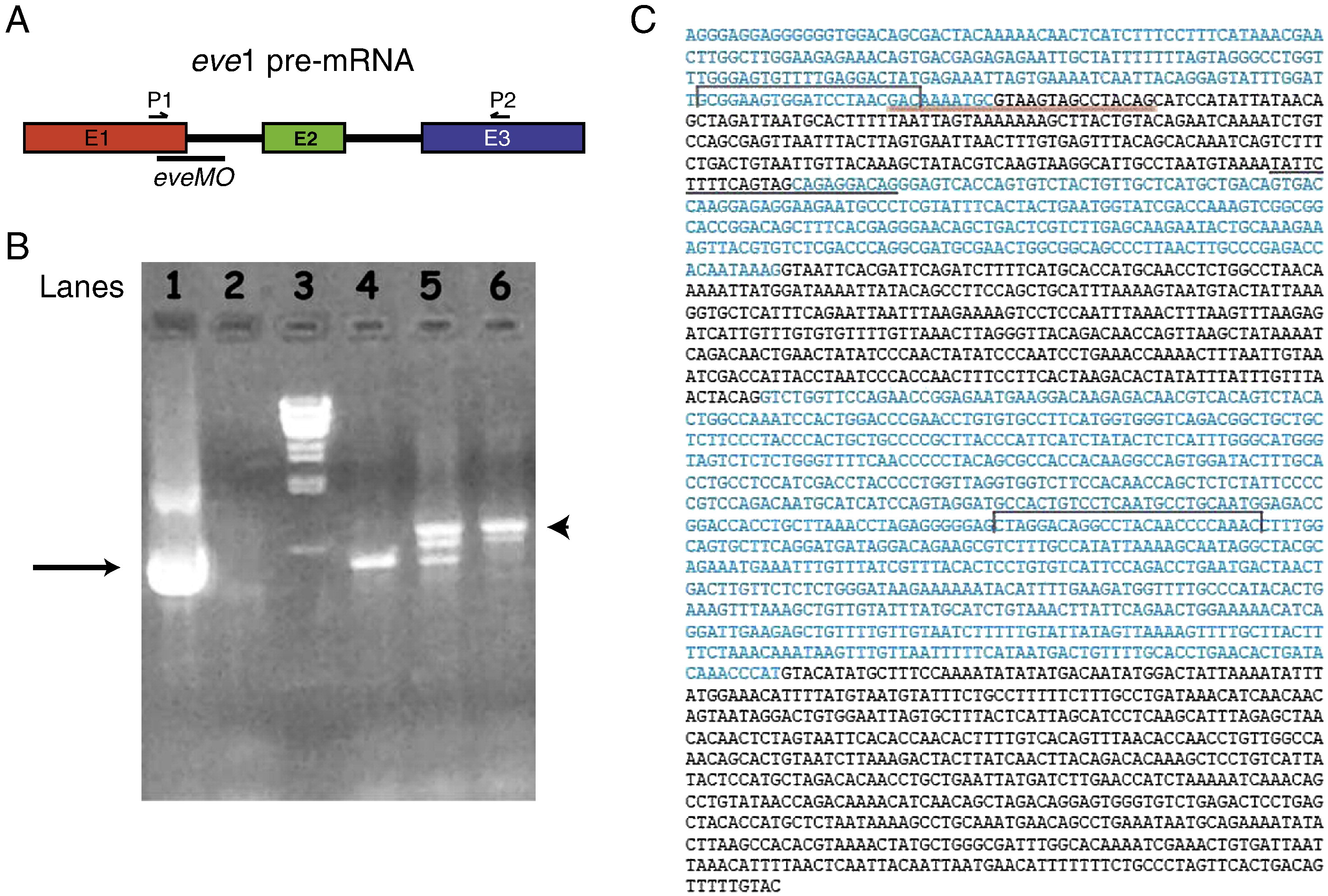
Fig. S1 Analysis of eve1 transcripts from uninjected and eveMO injected embryos at 6 hpf by RT-PCR. A) Schematic illustration of the eve1 pre-mRNA and the locations of designed eve1-specific splice blocking morpholino (eveMO) as well as the positions of primers used for the RT-PCR reactions. B) A 1.0% agarose gel with PCR products from various conditions. Lane 1) eve1 cDNA template used as positive control. Lane 2) No eve1 cDNA added as negative control. Lane 3) Lambda DNA markers, Lane 4) RT-template from uninjected embryos at 6 hpf. Lane 5) RT-template from embryos injected with 10 ng eveMO. Lane 6) RT-template from embryos injected with 25 ng eveMO. Black arrow indicates the expected normal PCR product of 674-bp. Arrowhead indicates the longer PCR product of 895-bp that contains the sequence of intron 1. C) Exon and intron sequences of the eve1 gene. Exon sequences are indicated in blue letters and intron sequences are indicated in black letters. Red line is used to indicate the location of eveMO blocking exon1 and intron 1 junction. Black line is used to indicate the location of eve1-MO blocking intron 1 and exon 2 junction. Black brackets indicate the location of primers that are used for RT-PCR experiments.
Reprinted from Developmental Biology, 349(1), Seebald, J.L., and Szeto, D.P., Zebrafish eve1 regulates the lateral and ventral fates of mesodermal progenitor cells at the onset of gastrulation, 78-89, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.