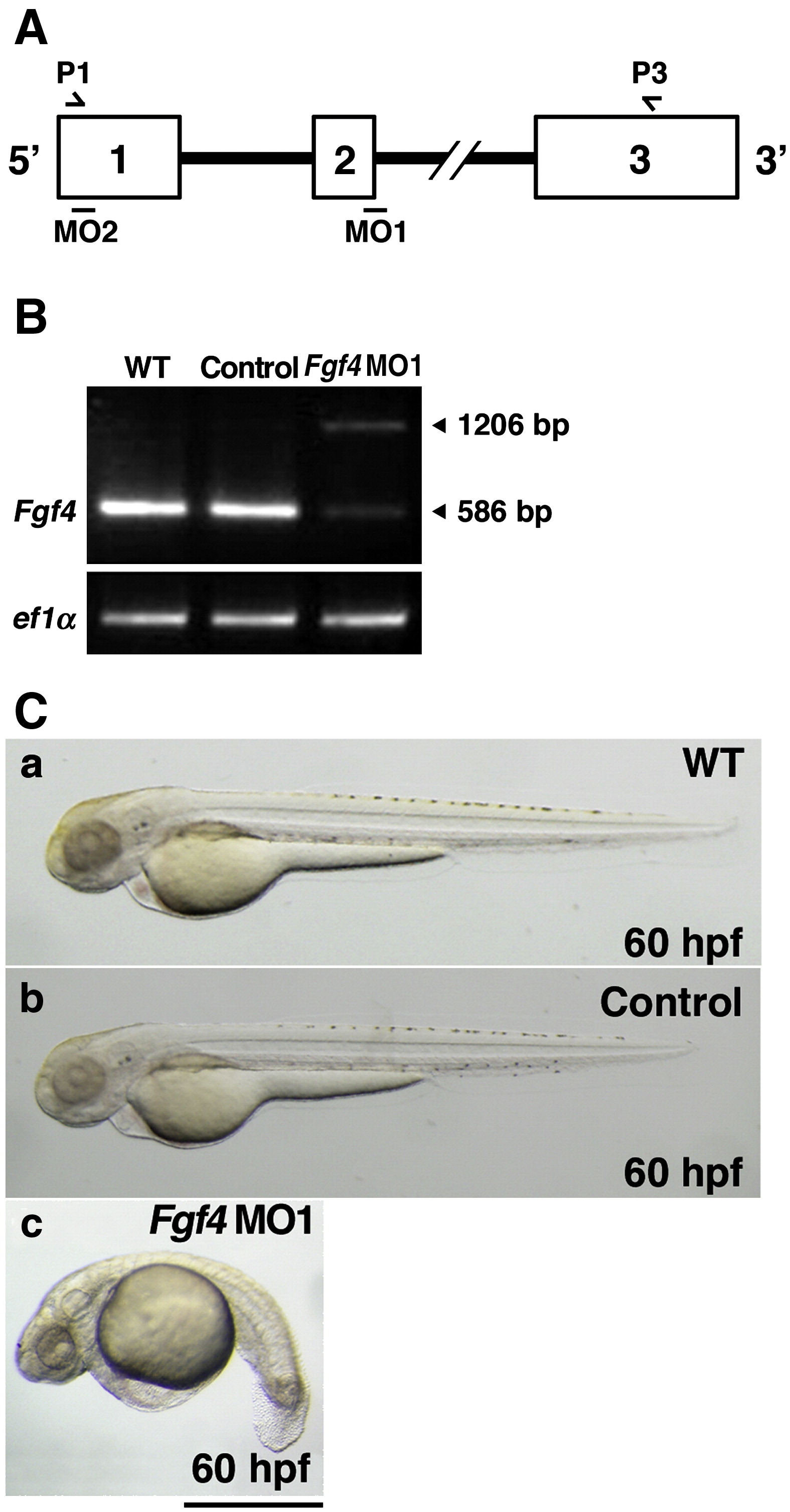
Fig. 1 Inhibition of Fgf4 functions in zebrafish embryos. (A) The coding region of zebrafish Fgf4 is divided by two introns. Open boxes and black lines indicate exons and introns, respectively. MO1 and MO2 indicate the target positions of Fgf4 MO1 and MO2, respectively. P1 and P3 indicate sites of P1 and P3 primers. (B) Zebrafish two-cell embryos were injected with Fgf4 MO1 (∼ 45 ng). The entire coding region of Fgf4 cDNA was amplified from wild-type or Fgf4 MO1-injected embryonic cDNA by RT-PCR using P1 and P3 primers. Arrowheads (586 bp and 1206 bp) indicate mature and unspliced Fgf4 cDNAs, respectively. Elongation factor 1-alpha (ef1α) cDNA was also amplified as a control. The cDNAs were analyzed by 1.2% agarose gel electrophoresis. After electrophoresis, the gel was stained with ethidium bromide. (C) Two-cell embryos were injected with standard control MO and Fgf4 MO1 (∼ 45 ng). Lateral views of the embryos at 60 hpf are shown. Control MO-injected embryos (97.2%, n = 36) (b) as well as the wild-type embryos (a) developed well. In contrast, Fgf4 MO-injected embryos showed bent trunks, pericardial edema and cerebral atrophy (67.1%, n = 73) (c). A scale bar = 500 μm.
Reprinted from Developmental Biology, 332(1), Yamauchi, H., Miyakawa, N., Miyake, A., and Itoh, N., Fgf4 is required for left-right patterning of visceral organs in zebrafish, 177-185, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.