- Title
-
Analysis of the senescence secretome during zebrafish retina regeneration
- Authors
- Konar, G.J., Vallone, K.T., Nguyen, T.D., Patton, J.G.
- Source
- Full text @ Front Aging
|
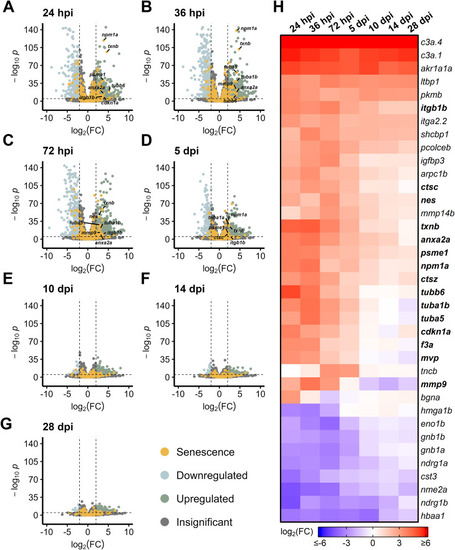
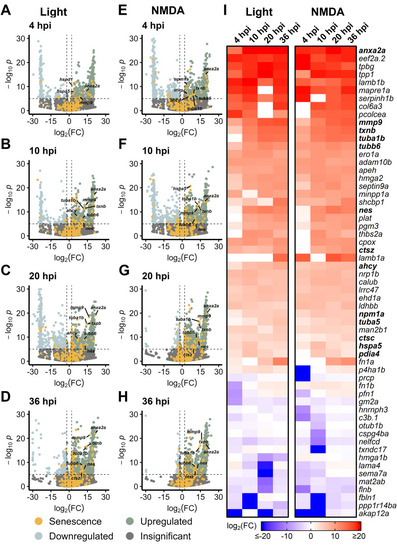
Differentially expressed SASP factors after acute light damage. Bulk RNAseq was performed on whole zebrafish retinas after acute light damage and compared to undamaged controls. |
|
Differentially expressed SASP factors after NMDA damage. Bulk RNAseq was performed on whole zebrafish retinas after intravitreal injections of NMDA and compared to undamaged controls. |
|
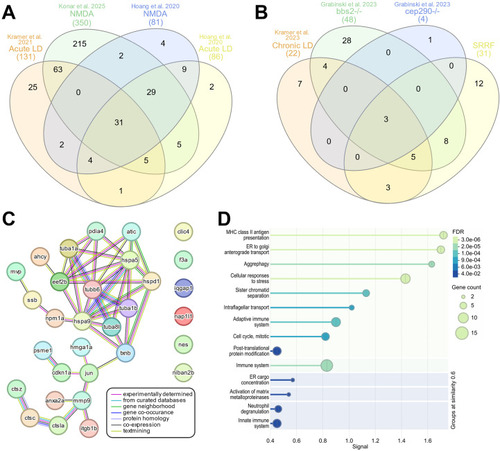
Gene Ontology (GO) analysis of differentially expressed SASP factors after light and NMDA damage. |
|
Upregulation of SASP factors in Müller glia after light and NMDA damage. Bulk RNAseq was performed on |
|
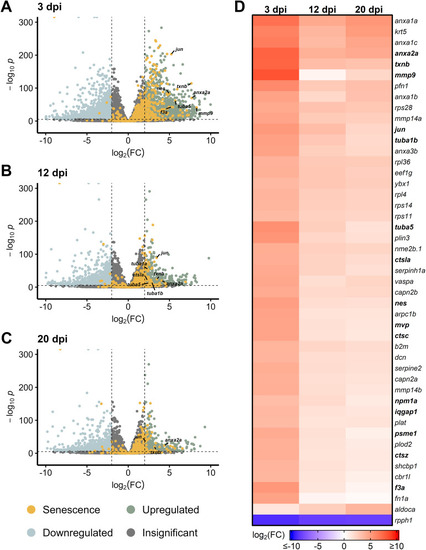
Differentially expressed SASP factors in genetic mutants with chronic photoreceptor damage. Bulk RNAseq was performed on whole zebrafish retinas from |
|
Regeneration specific secretome signature. InteractiVenn based analysis was used to determine the presence of a conserved secretome signature. |
|
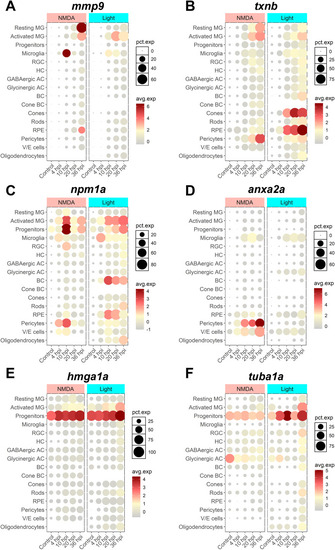
scRNAseq visualization of SASP factors after NMDA and light damage. Localization of SASP factor expression based on scRNAseq data from dissociated retinas was visualized using Seurat and the plot1cell package. |
|
Depletion of nucleophosmin 1a ( |