- Title
-
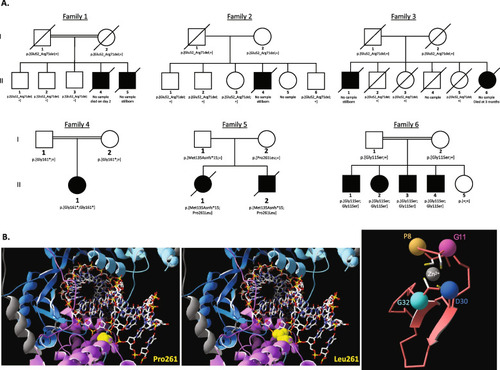
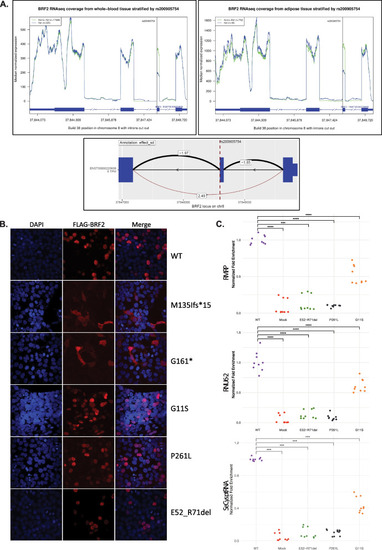
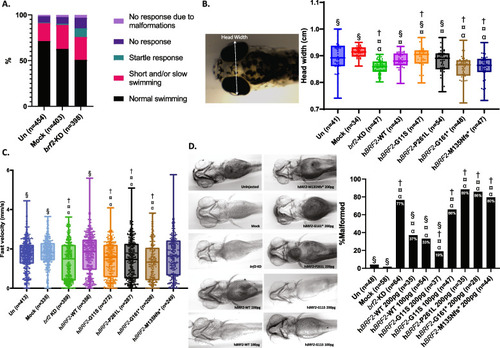
Bi-allelic variants in BRF2 are associated with perinatal death and craniofacial anomalies
- Authors
- Mattioli, F., Friðriksdóttir, R., Hebert, A., Bassani, S., Ibrahim, N., Naz, S., Chrast, J., Pailler-Pradeau, C., Oddsson, Á., Sulem, P., Halldorsson, G.H., Melsted, P., Guðbjartsson, D.F., Palombo, F., Pippucci, T., Nouri, N., Seri, M., Farrow, E.G., Saunders, C.J., Guex, N., Ansar, M., Stefansson, K., Reymond, A.
- Source
- Full text @ Genome Med.
|
Identified biallelic variants in |
|
Variants assessments. |
|
PHENOTYPE:
|