- Title
-
PDGFRA is a conserved HAND2 effector during early cardiac development
- Authors
- Xu, Y., Gehlot, R., Capon, S.J., Albu, M., Gretz, J., Bloomekatz, J., Mattonet, K., Vucicevic, D., Talyan, S., Kikhi, K., Günther, S., Looso, M., Firulli, B.A., Sanda, M., Firulli, A.B., Lacadie, S.A., Yelon, D., Stainier, D.Y.R.
- Source
- Full text @ Nat Cardiovasc Res
|
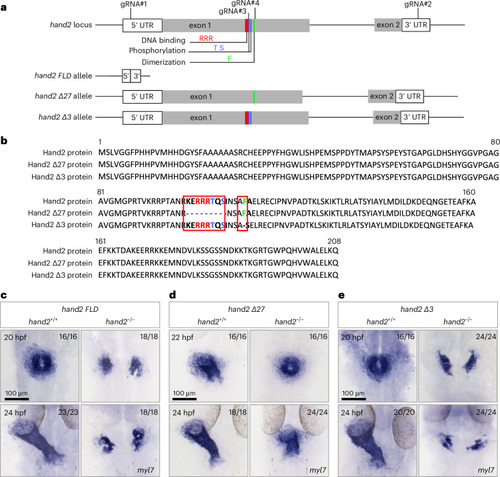
The DNA-binding domain of Hand2 is not required for early zebrafish cardiogenesis. |
|
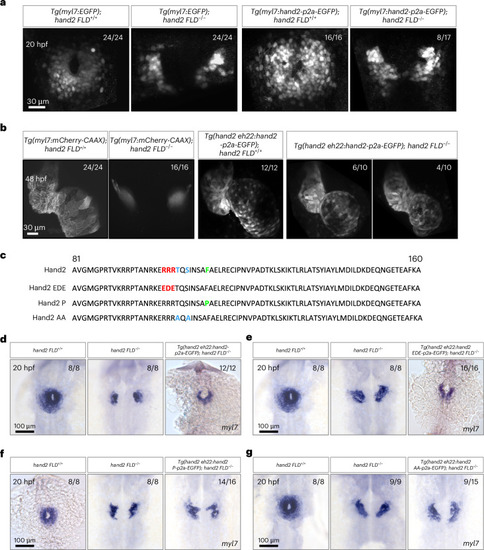
Cardiac fusion requires the dimerization domain of Hand2. |
|
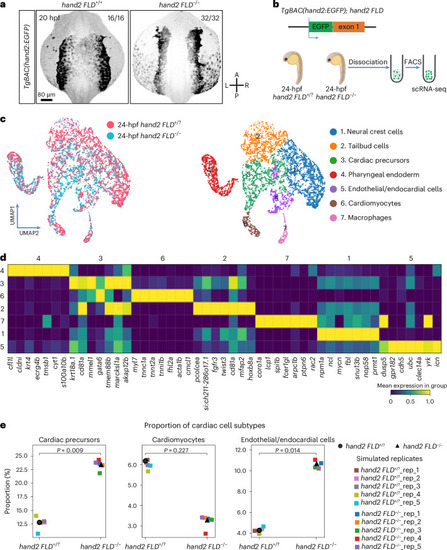
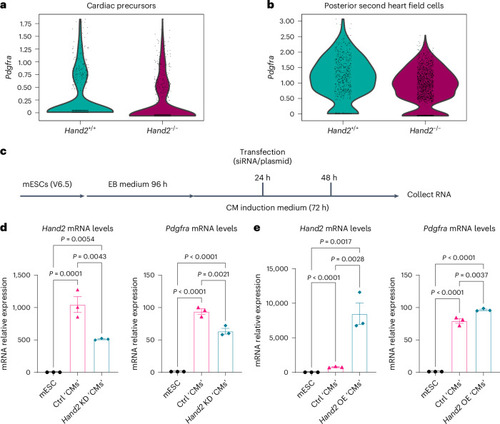
scRNA-seq analysis of |
|
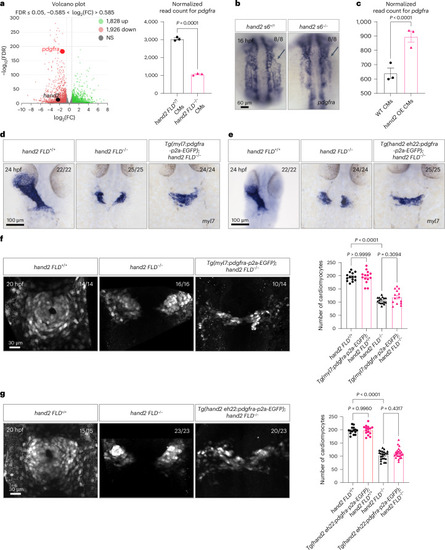
Hand2 regulates |
|
Deletion of a putative Hand2-binding region in a zebrafish |
|
|