- Title
-
The role of ATP-binding Cassette subfamily B member 6 in the inner ear
- Authors
- Baril, S.A., Wilson, K.A., Shaik, M.M., Fukuda, Y., Umans, R.A., Barbieri, A., Lynch, J., Gose, T., Myasnikov, A., Oldham, M.L., Wang, Y., Zhu, J., Fang, J., Zuo, J., Kalathur, R.C., Ford, R.C., Coffin, A., Taylor, M.R., O'Mara, M.L., Schuetz, J.D.
- Source
- Full text @ Nat. Commun.
|
ABCB6 variants identified in Dyschromatosis Universalis Hereditaria (DUH). |
|
Characterization of the L356P mutation. |
|
Characterization of zebrafish ABCB6 (Abcb6). |
|
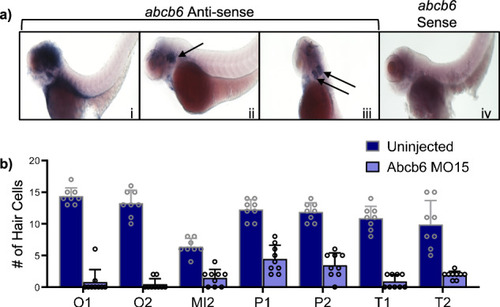
Zebrafish abcb6 knockdown affects lateral line development in zebrafish. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Zebrafish Abcb6 MO15 morpholino affects otolith development in zebrafish. PHENOTYPE:
|
|
Zebrafish Abcb6 MO15 morphants show altered behavior compared to uninjected zebrafish. Zebrafish movement paths overlaid with last video frame shows uninjected zebrafish swim smoothly in the upper 2/3 of the tank ( PHENOTYPE:
|
|
ABCB6 plays a role in the murine inner ear. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|